| Motif | BHE40.H13CORE.1.S.C |
| Gene (human) | BHLHE40 (GeneCards) |
| Gene synonyms (human) | BHLHB2, DEC1, SHARP2, STRA13 |
| Gene (mouse) | Bhlhe40 |
| Gene synonyms (mouse) | Bhlhb2, Clast5, Dec1, Stra13 |
| LOGO |  |
| LOGO (reverse complement) | 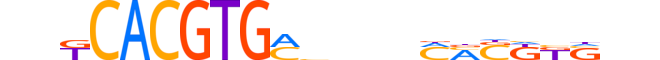 |
| Motif subtype | 1 |
| Quality | C |
| Motif | BHE40.H13CORE.1.S.C |
| Gene (human) | BHLHE40 (GeneCards) |
| Gene synonyms (human) | BHLHB2, DEC1, SHARP2, STRA13 |
| Gene (mouse) | Bhlhe40 |
| Gene synonyms (mouse) | Bhlhb2, Clast5, Dec1, Stra13 |
| LOGO |  |
| LOGO (reverse complement) | 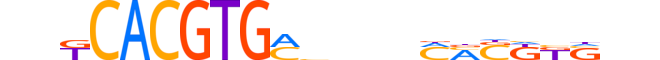 |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 22 |
| Consensus | nnSMMSKKnnnvKCACGTGMnn |
| GC content | 58.72% |
| Information content (bits; total / per base) | 17.51 / 0.796 |
| Data sources | HT-SELEX |
| Aligned words | 1866 |
| Previous names | BHE40.H12CORE.1.S.C |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (35) | 0.768 | 0.833 | 0.742 | 0.815 | 0.838 | 0.893 | 3.216 | 3.697 | 278.495 | 402.921 |
| Mouse | 7 (43) | 0.768 | 0.877 | 0.732 | 0.866 | 0.822 | 0.919 | 3.13 | 4.088 | 239.77 | 423.721 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 10 experiments | median | 0.997 | 0.995 | 0.977 | 0.973 | 0.851 | 0.862 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.998 | 0.997 | |
| Methyl HT-SELEX, 2 experiments | median | 0.99 | 0.983 | 0.949 | 0.94 | 0.814 | 0.82 |
| best | 0.993 | 0.988 | 0.957 | 0.951 | 0.824 | 0.83 | |
| Non-Methyl HT-SELEX, 8 experiments | median | 0.998 | 0.997 | 0.997 | 0.995 | 0.878 | 0.895 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.998 | 0.997 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 15.603 | 25.022 | 0.352 | 0.28 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | Hairy-related {1.2.4} (TFClass) |
| TF subfamily | HAIRY {1.2.4.1} (TFClass) |
| TFClass ID | TFClass: 1.2.4.1.12 |
| HGNC | HGNC:1046 |
| MGI | MGI:1097714 |
| EntrezGene (human) | GeneID:8553 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20893 (SSTAR profile) |
| UniProt ID (human) | BHE40_HUMAN |
| UniProt ID (mouse) | BHE40_MOUSE |
| UniProt AC (human) | O14503 (TFClass) |
| UniProt AC (mouse) | O35185 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 7 mouse |
| HT-SELEX | 8 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | BHE40.H13CORE.1.S.C.pcm |
| PWM | BHE40.H13CORE.1.S.C.pwm |
| PFM | BHE40.H13CORE.1.S.C.pfm |
| Threshold to P-value map | BHE40.H13CORE.1.S.C.thr |
| Motif in other formats | |
| JASPAR format | BHE40.H13CORE.1.S.C_jaspar_format.txt |
| MEME format | BHE40.H13CORE.1.S.C_meme_format.meme |
| Transfac format | BHE40.H13CORE.1.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 387.25 | 491.25 | 554.25 | 433.25 |
| 02 | 384.75 | 270.75 | 645.75 | 564.75 |
| 03 | 176.0 | 1276.0 | 230.0 | 184.0 |
| 04 | 1196.0 | 298.0 | 256.0 | 116.0 |
| 05 | 294.0 | 1281.0 | 139.0 | 152.0 |
| 06 | 141.0 | 295.0 | 1326.0 | 104.0 |
| 07 | 147.0 | 210.0 | 305.0 | 1204.0 |
| 08 | 136.0 | 117.0 | 1274.0 | 339.0 |
| 09 | 543.0 | 469.0 | 495.0 | 359.0 |
| 10 | 456.0 | 471.0 | 500.0 | 439.0 |
| 11 | 479.0 | 349.0 | 659.0 | 379.0 |
| 12 | 524.0 | 351.0 | 726.0 | 265.0 |
| 13 | 25.0 | 13.0 | 916.0 | 912.0 |
| 14 | 0.0 | 1865.0 | 1.0 | 0.0 |
| 15 | 1866.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 1866.0 | 0.0 | 0.0 |
| 17 | 0.0 | 0.0 | 1866.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 1866.0 |
| 19 | 0.0 | 0.0 | 1866.0 | 0.0 |
| 20 | 1051.0 | 706.0 | 42.0 | 67.0 |
| 21 | 295.25 | 572.25 | 429.25 | 569.25 |
| 22 | 429.25 | 518.25 | 454.25 | 464.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.208 | 0.263 | 0.297 | 0.232 |
| 02 | 0.206 | 0.145 | 0.346 | 0.303 |
| 03 | 0.094 | 0.684 | 0.123 | 0.099 |
| 04 | 0.641 | 0.16 | 0.137 | 0.062 |
| 05 | 0.158 | 0.686 | 0.074 | 0.081 |
| 06 | 0.076 | 0.158 | 0.711 | 0.056 |
| 07 | 0.079 | 0.113 | 0.163 | 0.645 |
| 08 | 0.073 | 0.063 | 0.683 | 0.182 |
| 09 | 0.291 | 0.251 | 0.265 | 0.192 |
| 10 | 0.244 | 0.252 | 0.268 | 0.235 |
| 11 | 0.257 | 0.187 | 0.353 | 0.203 |
| 12 | 0.281 | 0.188 | 0.389 | 0.142 |
| 13 | 0.013 | 0.007 | 0.491 | 0.489 |
| 14 | 0.0 | 0.999 | 0.001 | 0.0 |
| 15 | 1.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 1.0 | 0.0 | 0.0 |
| 17 | 0.0 | 0.0 | 1.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 1.0 |
| 19 | 0.0 | 0.0 | 1.0 | 0.0 |
| 20 | 0.563 | 0.378 | 0.023 | 0.036 |
| 21 | 0.158 | 0.307 | 0.23 | 0.305 |
| 22 | 0.23 | 0.278 | 0.243 | 0.249 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.185 | 0.051 | 0.172 | -0.074 |
| 02 | -0.192 | -0.541 | 0.324 | 0.19 |
| 03 | -0.968 | 1.004 | -0.703 | -0.924 |
| 04 | 0.939 | -0.446 | -0.597 | -1.38 |
| 05 | -0.459 | 1.008 | -1.201 | -1.113 |
| 06 | -1.187 | -0.456 | 1.042 | -1.487 |
| 07 | -1.146 | -0.793 | -0.423 | 0.946 |
| 08 | -1.223 | -1.371 | 1.002 | -0.318 |
| 09 | 0.151 | 0.005 | 0.059 | -0.261 |
| 10 | -0.023 | 0.01 | 0.069 | -0.061 |
| 11 | 0.026 | -0.289 | 0.344 | -0.207 |
| 12 | 0.116 | -0.283 | 0.441 | -0.562 |
| 13 | -2.858 | -3.449 | 0.673 | 0.668 |
| 14 | -5.516 | 1.383 | -5.09 | -5.516 |
| 15 | 1.383 | -5.516 | -5.516 | -5.516 |
| 16 | -5.516 | 1.383 | -5.516 | -5.516 |
| 17 | -5.516 | -5.516 | 1.383 | -5.516 |
| 18 | -5.516 | -5.516 | -5.516 | 1.383 |
| 19 | -5.516 | -5.516 | 1.383 | -5.516 |
| 20 | 0.81 | 0.413 | -2.368 | -1.917 |
| 21 | -0.455 | 0.204 | -0.083 | 0.198 |
| 22 | -0.083 | 0.105 | -0.027 | -0.005 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.19554 |
| 0.0005 | 1.46671 |
| 0.0001 | 4.99781 |