| Motif | CEBPE.H13RSNP.0.P.B |
| Gene (human) | CEBPE (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Cebpe |
| Gene synonyms (mouse) | Gm294 |
| LOGO |  |
| LOGO (reverse complement) | 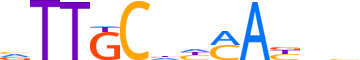 |
| Motif subtype | 0 |
| Quality | B |
| Motif | CEBPE.H13RSNP.0.P.B |
| Gene (human) | CEBPE (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Cebpe |
| Gene synonyms (mouse) | Gm294 |
| LOGO |  |
| LOGO (reverse complement) | 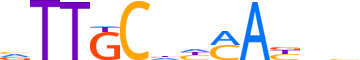 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 12 |
| Consensus | vdvTKddGCAAb |
| GC content | 46.82% |
| Information content (bits; total / per base) | 11.353 / 0.946 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | CEBPE.H12RSNP.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 3 (21) | 0.924 | 0.935 | 0.842 | 0.87 | 0.865 | 0.874 | 2.614 | 2.687 | 262.62 | 335.921 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.974 | 0.952 | 0.954 | 0.933 | 0.85 | 0.846 |
| best | 0.993 | 0.988 | 0.993 | 0.986 | 0.992 | 0.985 | |
| Methyl HT-SELEX, 2 experiments | median | 0.953 | 0.916 | 0.928 | 0.893 | 0.837 | 0.822 |
| best | 0.966 | 0.938 | 0.963 | 0.934 | 0.933 | 0.904 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.984 | 0.971 | 0.965 | 0.951 | 0.872 | 0.876 |
| best | 0.993 | 0.988 | 0.993 | 0.986 | 0.992 | 0.985 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.916 | 0.797 | 0.906 | 0.645 |
| batch 2 | 0.87 | 0.731 | 0.877 | 0.676 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | C/EBP-related {1.1.8} (TFClass) |
| TF subfamily | CEBP {1.1.8.1} (TFClass) |
| TFClass ID | TFClass: 1.1.8.1.5 |
| HGNC | HGNC:1836 |
| MGI | MGI:103572 |
| EntrezGene (human) | GeneID:1053 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:110794 (SSTAR profile) |
| UniProt ID (human) | CEBPE_HUMAN |
| UniProt ID (mouse) | CEBPE_MOUSE |
| UniProt AC (human) | Q15744 (TFClass) |
| UniProt AC (mouse) | Q6PZD9 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 3 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | CEBPE.H13RSNP.0.P.B.pcm |
| PWM | CEBPE.H13RSNP.0.P.B.pwm |
| PFM | CEBPE.H13RSNP.0.P.B.pfm |
| Threshold to P-value map | CEBPE.H13RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | CEBPE.H13RSNP.0.P.B_jaspar_format.txt |
| MEME format | CEBPE.H13RSNP.0.P.B_meme_format.meme |
| Transfac format | CEBPE.H13RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 225.0 | 284.0 | 372.0 | 119.0 |
| 02 | 253.0 | 109.0 | 331.0 | 307.0 |
| 03 | 293.0 | 241.0 | 456.0 | 10.0 |
| 04 | 5.0 | 1.0 | 1.0 | 993.0 |
| 05 | 3.0 | 1.0 | 507.0 | 489.0 |
| 06 | 266.0 | 19.0 | 475.0 | 240.0 |
| 07 | 219.0 | 100.0 | 268.0 | 413.0 |
| 08 | 1.0 | 1.0 | 996.0 | 2.0 |
| 09 | 154.0 | 808.0 | 1.0 | 37.0 |
| 10 | 993.0 | 7.0 | 0.0 | 0.0 |
| 11 | 1000.0 | 0.0 | 0.0 | 0.0 |
| 12 | 31.0 | 368.0 | 272.0 | 329.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.225 | 0.284 | 0.372 | 0.119 |
| 02 | 0.253 | 0.109 | 0.331 | 0.307 |
| 03 | 0.293 | 0.241 | 0.456 | 0.01 |
| 04 | 0.005 | 0.001 | 0.001 | 0.993 |
| 05 | 0.003 | 0.001 | 0.507 | 0.489 |
| 06 | 0.266 | 0.019 | 0.475 | 0.24 |
| 07 | 0.219 | 0.1 | 0.268 | 0.413 |
| 08 | 0.001 | 0.001 | 0.996 | 0.002 |
| 09 | 0.154 | 0.808 | 0.001 | 0.037 |
| 10 | 0.993 | 0.007 | 0.0 | 0.0 |
| 11 | 1.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.031 | 0.368 | 0.272 | 0.329 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.105 | 0.127 | 0.395 | -0.735 |
| 02 | 0.012 | -0.821 | 0.279 | 0.204 |
| 03 | 0.158 | -0.036 | 0.598 | -3.066 |
| 04 | -3.622 | -4.525 | -4.525 | 1.374 |
| 05 | -3.975 | -4.525 | 0.704 | 0.668 |
| 06 | 0.062 | -2.497 | 0.639 | -0.041 |
| 07 | -0.131 | -0.906 | 0.069 | 0.499 |
| 08 | -4.525 | -4.525 | 1.377 | -4.213 |
| 09 | -0.48 | 1.168 | -4.525 | -1.872 |
| 10 | 1.374 | -3.362 | -4.982 | -4.982 |
| 11 | 1.381 | -4.982 | -4.982 | -4.982 |
| 12 | -2.04 | 0.384 | 0.084 | 0.273 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.73677 |
| 0.0005 | 5.374975 |
| 0.0001 | 7.8941 |