| Motif | GATA1.H13RSNP.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO | 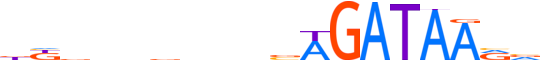 |
| LOGO (reverse complement) | 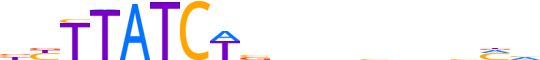 |
| Motif subtype | 0 |
| Quality | B |
| Motif | GATA1.H13RSNP.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO | 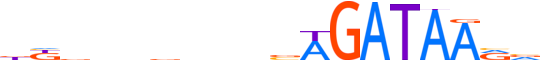 |
| LOGO (reverse complement) | 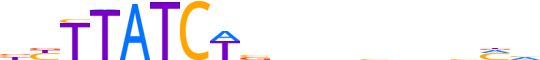 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 18 |
| Consensus | nKbnndnnnvWGATAAvv |
| GC content | 43.79% |
| Information content (bits; total / per base) | 11.68 / 0.649 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | GATA1.H12RSNP.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 21 (139) | 0.937 | 0.977 | 0.874 | 0.955 | 0.926 | 0.961 | 3.782 | 4.419 | 297.796 | 408.886 |
| Mouse | 37 (221) | 0.893 | 0.956 | 0.799 | 0.907 | 0.875 | 0.936 | 3.21 | 3.868 | 182.569 | 402.638 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.884 | 0.844 | 0.83 | 0.794 | 0.755 | 0.732 |
| best | 0.958 | 0.928 | 0.938 | 0.904 | 0.863 | 0.836 | |
| Methyl HT-SELEX, 1 experiments | median | 0.958 | 0.928 | 0.938 | 0.904 | 0.863 | 0.836 |
| best | 0.958 | 0.928 | 0.938 | 0.904 | 0.863 | 0.836 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.811 | 0.759 | 0.723 | 0.685 | 0.647 | 0.629 |
| best | 0.811 | 0.759 | 0.723 | 0.685 | 0.647 | 0.629 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 4.184 | 13.07 | 0.289 | 0.191 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.1 |
| HGNC | HGNC:4170 |
| MGI | MGI:95661 |
| EntrezGene (human) | GeneID:2623 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14460 (SSTAR profile) |
| UniProt ID (human) | GATA1_HUMAN |
| UniProt ID (mouse) | GATA1_MOUSE |
| UniProt AC (human) | P15976 (TFClass) |
| UniProt AC (mouse) | P17679 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 21 human, 37 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | GATA1.H13RSNP.0.P.B.pcm |
| PWM | GATA1.H13RSNP.0.P.B.pwm |
| PFM | GATA1.H13RSNP.0.P.B.pfm |
| Threshold to P-value map | GATA1.H13RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA1.H13RSNP.0.P.B_jaspar_format.txt |
| MEME format | GATA1.H13RSNP.0.P.B_meme_format.meme |
| Transfac format | GATA1.H13RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 142.0 | 142.0 | 223.0 | 493.0 |
| 02 | 92.0 | 95.0 | 584.0 | 229.0 |
| 03 | 139.0 | 241.0 | 407.0 | 213.0 |
| 04 | 233.0 | 199.0 | 279.0 | 289.0 |
| 05 | 304.0 | 193.0 | 295.0 | 208.0 |
| 06 | 253.0 | 156.0 | 402.0 | 189.0 |
| 07 | 242.0 | 210.0 | 327.0 | 221.0 |
| 08 | 210.0 | 227.0 | 291.0 | 272.0 |
| 09 | 298.0 | 182.0 | 315.0 | 205.0 |
| 10 | 159.0 | 442.0 | 291.0 | 108.0 |
| 11 | 653.25 | 8.25 | 7.25 | 331.25 |
| 12 | 0.25 | 1.25 | 998.25 | 0.25 |
| 13 | 993.25 | 0.25 | 2.25 | 4.25 |
| 14 | 0.25 | 6.25 | 5.25 | 988.25 |
| 15 | 963.25 | 0.25 | 13.25 | 23.25 |
| 16 | 890.25 | 6.25 | 85.25 | 18.25 |
| 17 | 211.25 | 127.25 | 581.25 | 80.25 |
| 18 | 388.25 | 174.25 | 365.25 | 72.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.142 | 0.142 | 0.223 | 0.493 |
| 02 | 0.092 | 0.095 | 0.584 | 0.229 |
| 03 | 0.139 | 0.241 | 0.407 | 0.213 |
| 04 | 0.233 | 0.199 | 0.279 | 0.289 |
| 05 | 0.304 | 0.193 | 0.295 | 0.208 |
| 06 | 0.253 | 0.156 | 0.402 | 0.189 |
| 07 | 0.242 | 0.21 | 0.327 | 0.221 |
| 08 | 0.21 | 0.227 | 0.291 | 0.272 |
| 09 | 0.298 | 0.182 | 0.315 | 0.205 |
| 10 | 0.159 | 0.442 | 0.291 | 0.108 |
| 11 | 0.653 | 0.008 | 0.007 | 0.331 |
| 12 | 0.0 | 0.001 | 0.998 | 0.0 |
| 13 | 0.993 | 0.0 | 0.002 | 0.004 |
| 14 | 0.0 | 0.006 | 0.005 | 0.988 |
| 15 | 0.963 | 0.0 | 0.013 | 0.023 |
| 16 | 0.89 | 0.006 | 0.085 | 0.018 |
| 17 | 0.211 | 0.127 | 0.581 | 0.08 |
| 18 | 0.388 | 0.174 | 0.365 | 0.072 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.56 | -0.56 | -0.113 | 0.676 |
| 02 | -0.988 | -0.956 | 0.845 | -0.087 |
| 03 | -0.582 | -0.036 | 0.485 | -0.159 |
| 04 | -0.07 | -0.226 | 0.109 | 0.144 |
| 05 | 0.194 | -0.257 | 0.164 | -0.183 |
| 06 | 0.012 | -0.467 | 0.472 | -0.278 |
| 07 | -0.032 | -0.173 | 0.267 | -0.122 |
| 08 | -0.173 | -0.096 | 0.151 | 0.084 |
| 09 | 0.175 | -0.315 | 0.23 | -0.197 |
| 10 | -0.449 | 0.567 | 0.151 | -0.83 |
| 11 | 0.956 | -3.228 | -3.334 | 0.28 |
| 12 | -4.847 | -4.437 | 1.379 | -4.847 |
| 13 | 1.374 | -4.847 | -4.148 | -3.74 |
| 14 | -4.847 | -3.452 | -3.586 | 1.369 |
| 15 | 1.344 | -4.847 | -2.822 | -2.31 |
| 16 | 1.265 | -3.452 | -1.063 | -2.534 |
| 17 | -0.167 | -0.669 | 0.84 | -1.122 |
| 18 | 0.438 | -0.358 | 0.377 | -1.225 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.08826 |
| 0.0005 | 5.25941 |
| 0.0001 | 7.51601 |