| Motif | GCR.H13CORE.0.PS.A |
| Gene (human) | NR3C1 (GeneCards) |
| Gene synonyms (human) | GRL |
| Gene (mouse) | Nr3c1 |
| Gene synonyms (mouse) | Grl, Grl1 |
| LOGO |  |
| LOGO (reverse complement) | 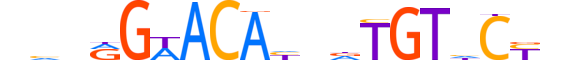 |
| Motif subtype | 0 |
| Quality | A |
| Motif | GCR.H13CORE.0.PS.A |
| Gene (human) | NR3C1 (GeneCards) |
| Gene synonyms (human) | GRL |
| Gene (mouse) | Nr3c1 |
| Gene synonyms (mouse) | Grl, Grl1 |
| LOGO |  |
| LOGO (reverse complement) | 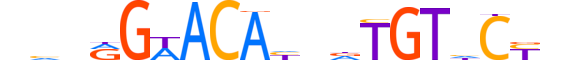 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 19 |
| Consensus | nRGdACAhndTGTWCYndn |
| GC content | 44.1% |
| Information content (bits; total / per base) | 17.293 / 0.91 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 1686 |
| Previous names | GCR.H12CORE.0.PS.A |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 82 (479) | 0.869 | 0.963 | 0.814 | 0.927 | 0.833 | 0.94 | 3.343 | 4.82 | 201.137 | 822.538 |
| Mouse | 93 (512) | 0.833 | 0.988 | 0.772 | 0.977 | 0.794 | 0.985 | 3.119 | 5.959 | 192.27 | 719.284 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.767 | 0.776 | 0.638 | 0.652 | 0.575 | 0.591 |
| best | 0.838 | 0.848 | 0.672 | 0.692 | 0.592 | 0.615 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 2.776 | 17.998 | 0.167 | 0.069 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Steroid hormone receptors {2.1.1} (TFClass) |
| TF subfamily | GR-like (NR3C) {2.1.1.1} (TFClass) |
| TFClass ID | TFClass: 2.1.1.1.1 |
| HGNC | HGNC:7978 |
| MGI | MGI:95824 |
| EntrezGene (human) | GeneID:2908 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | GCR_HUMAN |
| UniProt ID (mouse) | GCR_MOUSE |
| UniProt AC (human) | P04150 (TFClass) |
| UniProt AC (mouse) | P06537 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 82 human, 93 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | GCR.H13CORE.0.PS.A.pcm |
| PWM | GCR.H13CORE.0.PS.A.pwm |
| PFM | GCR.H13CORE.0.PS.A.pfm |
| Threshold to P-value map | GCR.H13CORE.0.PS.A.thr |
| Motif in other formats | |
| JASPAR format | GCR.H13CORE.0.PS.A_jaspar_format.txt |
| MEME format | GCR.H13CORE.0.PS.A_meme_format.meme |
| Transfac format | GCR.H13CORE.0.PS.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 424.75 | 455.75 | 418.75 | 386.75 |
| 02 | 1012.75 | 176.75 | 465.75 | 30.75 |
| 03 | 64.0 | 18.0 | 1579.0 | 25.0 |
| 04 | 648.0 | 139.0 | 337.0 | 562.0 |
| 05 | 1637.0 | 10.0 | 25.0 | 14.0 |
| 06 | 11.0 | 1646.0 | 12.0 | 17.0 |
| 07 | 1496.0 | 11.0 | 150.0 | 29.0 |
| 08 | 233.0 | 529.0 | 120.0 | 804.0 |
| 09 | 583.0 | 336.0 | 323.0 | 444.0 |
| 10 | 881.0 | 147.0 | 471.0 | 187.0 |
| 11 | 30.0 | 39.0 | 8.0 | 1609.0 |
| 12 | 0.0 | 0.0 | 1686.0 | 0.0 |
| 13 | 8.0 | 0.0 | 0.0 | 1678.0 |
| 14 | 796.0 | 49.0 | 6.0 | 835.0 |
| 15 | 0.0 | 1686.0 | 0.0 | 0.0 |
| 16 | 148.0 | 1002.0 | 40.0 | 496.0 |
| 17 | 307.0 | 536.0 | 288.0 | 555.0 |
| 18 | 319.5 | 166.5 | 482.5 | 717.5 |
| 19 | 373.75 | 318.75 | 447.75 | 545.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.252 | 0.27 | 0.248 | 0.229 |
| 02 | 0.601 | 0.105 | 0.276 | 0.018 |
| 03 | 0.038 | 0.011 | 0.937 | 0.015 |
| 04 | 0.384 | 0.082 | 0.2 | 0.333 |
| 05 | 0.971 | 0.006 | 0.015 | 0.008 |
| 06 | 0.007 | 0.976 | 0.007 | 0.01 |
| 07 | 0.887 | 0.007 | 0.089 | 0.017 |
| 08 | 0.138 | 0.314 | 0.071 | 0.477 |
| 09 | 0.346 | 0.199 | 0.192 | 0.263 |
| 10 | 0.523 | 0.087 | 0.279 | 0.111 |
| 11 | 0.018 | 0.023 | 0.005 | 0.954 |
| 12 | 0.0 | 0.0 | 1.0 | 0.0 |
| 13 | 0.005 | 0.0 | 0.0 | 0.995 |
| 14 | 0.472 | 0.029 | 0.004 | 0.495 |
| 15 | 0.0 | 1.0 | 0.0 | 0.0 |
| 16 | 0.088 | 0.594 | 0.024 | 0.294 |
| 17 | 0.182 | 0.318 | 0.171 | 0.329 |
| 18 | 0.19 | 0.099 | 0.286 | 0.426 |
| 19 | 0.222 | 0.189 | 0.266 | 0.324 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.008 | 0.078 | -0.007 | -0.086 |
| 02 | 0.874 | -0.863 | 0.099 | -2.564 |
| 03 | -1.861 | -3.06 | 1.318 | -2.758 |
| 04 | 0.429 | -1.1 | -0.223 | 0.287 |
| 05 | 1.354 | -3.575 | -2.758 | -3.285 |
| 06 | -3.494 | 1.359 | -3.419 | -3.111 |
| 07 | 1.264 | -3.494 | -1.025 | -2.619 |
| 08 | -0.589 | 0.226 | -1.245 | 0.644 |
| 09 | 0.323 | -0.226 | -0.265 | 0.052 |
| 10 | 0.735 | -1.045 | 0.111 | -0.807 |
| 11 | -2.587 | -2.338 | -3.76 | 1.336 |
| 12 | -5.429 | -5.429 | 1.383 | -5.429 |
| 13 | -3.76 | -5.429 | -5.429 | 1.378 |
| 14 | 0.634 | -2.119 | -3.987 | 0.681 |
| 15 | -5.429 | 1.383 | -5.429 | -5.429 |
| 16 | -1.039 | 0.863 | -2.314 | 0.162 |
| 17 | -0.315 | 0.239 | -0.379 | 0.274 |
| 18 | -0.276 | -0.922 | 0.135 | 0.53 |
| 19 | -0.12 | -0.278 | 0.06 | 0.257 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.84911 |
| 0.0005 | 2.34446 |
| 0.0001 | 5.45991 |