| Motif | GMEB1.H13RSNP.1.P.C |
| Gene (human) | GMEB1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gmeb1 |
| Gene synonyms (mouse) | |
| LOGO | 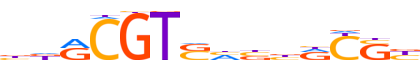 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | C |
| Motif | GMEB1.H13RSNP.1.P.C |
| Gene (human) | GMEB1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gmeb1 |
| Gene synonyms (mouse) | |
| LOGO | 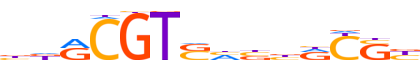 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 14 |
| Consensus | bdRCGTShbbRYSb |
| GC content | 65.57% |
| Information content (bits; total / per base) | 10.011 / 0.715 |
| Data sources | ChIP-Seq |
| Aligned words | 996 |
| Previous names | GMEB1.H12RSNP.1.P.C |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.845 | 0.867 | 0.753 | 0.775 | 0.751 | 0.777 | 2.511 | 2.657 | 84.824 | 115.119 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.796 | 0.763 | 0.751 | 0.719 | 0.693 | 0.67 |
| best | 0.957 | 0.934 | 0.899 | 0.869 | 0.813 | 0.788 | |
| Methyl HT-SELEX, 1 experiments | median | 0.636 | 0.593 | 0.603 | 0.57 | 0.574 | 0.553 |
| best | 0.636 | 0.593 | 0.603 | 0.57 | 0.574 | 0.553 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.957 | 0.934 | 0.899 | 0.869 | 0.813 | 0.788 |
| best | 0.957 | 0.934 | 0.899 | 0.869 | 0.813 | 0.788 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.701 | 0.079 | 0.275 | 0.333 |
| TF superclass | alpha-Helices exposed by beta-structures {5} (TFClass) |
| TF class | SAND domain factors {5.3} (TFClass) |
| TF family | GMEB {5.3.3} (TFClass) |
| TF subfamily | GMEB {5.3.3.1} (TFClass) |
| TFClass ID | TFClass: 5.3.3.1.1 |
| HGNC | HGNC:4370 |
| MGI | MGI:2135604 |
| EntrezGene (human) | GeneID:10691 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:56809 (SSTAR profile) |
| UniProt ID (human) | GMEB1_HUMAN |
| UniProt ID (mouse) | GMEB1_MOUSE |
| UniProt AC (human) | Q9Y692 (TFClass) |
| UniProt AC (mouse) | Q9JL60 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | GMEB1.H13RSNP.1.P.C.pcm |
| PWM | GMEB1.H13RSNP.1.P.C.pwm |
| PFM | GMEB1.H13RSNP.1.P.C.pfm |
| Threshold to P-value map | GMEB1.H13RSNP.1.P.C.thr |
| Motif in other formats | |
| JASPAR format | GMEB1.H13RSNP.1.P.C_jaspar_format.txt |
| MEME format | GMEB1.H13RSNP.1.P.C_meme_format.meme |
| Transfac format | GMEB1.H13RSNP.1.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 75.0 | 340.0 | 254.0 | 327.0 |
| 02 | 155.0 | 74.0 | 304.0 | 463.0 |
| 03 | 393.0 | 24.0 | 553.0 | 26.0 |
| 04 | 38.0 | 915.0 | 6.0 | 37.0 |
| 05 | 6.0 | 22.0 | 946.0 | 22.0 |
| 06 | 10.0 | 14.0 | 23.0 | 949.0 |
| 07 | 78.0 | 510.0 | 386.0 | 22.0 |
| 08 | 499.0 | 302.0 | 70.0 | 125.0 |
| 09 | 31.0 | 384.0 | 415.0 | 166.0 |
| 10 | 75.0 | 445.0 | 165.0 | 311.0 |
| 11 | 200.0 | 79.0 | 598.0 | 119.0 |
| 12 | 52.0 | 797.0 | 40.0 | 107.0 |
| 13 | 26.0 | 164.0 | 706.0 | 100.0 |
| 14 | 28.0 | 453.0 | 154.0 | 361.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.075 | 0.341 | 0.255 | 0.328 |
| 02 | 0.156 | 0.074 | 0.305 | 0.465 |
| 03 | 0.395 | 0.024 | 0.555 | 0.026 |
| 04 | 0.038 | 0.919 | 0.006 | 0.037 |
| 05 | 0.006 | 0.022 | 0.95 | 0.022 |
| 06 | 0.01 | 0.014 | 0.023 | 0.953 |
| 07 | 0.078 | 0.512 | 0.388 | 0.022 |
| 08 | 0.501 | 0.303 | 0.07 | 0.126 |
| 09 | 0.031 | 0.386 | 0.417 | 0.167 |
| 10 | 0.075 | 0.447 | 0.166 | 0.312 |
| 11 | 0.201 | 0.079 | 0.6 | 0.119 |
| 12 | 0.052 | 0.8 | 0.04 | 0.107 |
| 13 | 0.026 | 0.165 | 0.709 | 0.1 |
| 14 | 0.028 | 0.455 | 0.155 | 0.362 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.184 | 0.31 | 0.02 | 0.271 |
| 02 | -0.47 | -1.197 | 0.198 | 0.617 |
| 03 | 0.454 | -2.277 | 0.794 | -2.202 |
| 04 | -1.842 | 1.296 | -3.48 | -1.868 |
| 05 | -3.48 | -2.358 | 1.33 | -2.358 |
| 06 | -3.063 | -2.769 | -2.317 | 1.333 |
| 07 | -1.146 | 0.713 | 0.436 | -2.358 |
| 08 | 0.692 | 0.192 | -1.252 | -0.682 |
| 09 | -2.036 | 0.431 | 0.508 | -0.402 |
| 10 | -1.184 | 0.578 | -0.408 | 0.221 |
| 11 | -0.217 | -1.133 | 0.872 | -0.731 |
| 12 | -1.54 | 1.159 | -1.793 | -0.836 |
| 13 | -2.202 | -0.414 | 1.038 | -0.902 |
| 14 | -2.132 | 0.595 | -0.476 | 0.369 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.72011 |
| 0.0005 | 5.55711 |
| 0.0001 | 7.26561 |