| Motif | HMG20A.H13CORE.0.SB.A |
| Gene (human) | HMG20A (GeneCards) |
| Gene synonyms (human) | HMGX1, HMGXB1 |
| Gene (mouse) | Hmg20a |
| Gene synonyms (mouse) | Ibraf |
| LOGO |  |
| LOGO (reverse complement) | 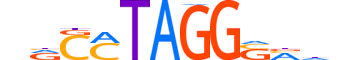 |
| Motif subtype | 0 |
| Quality | A |
| Motif | HMG20A.H13CORE.0.SB.A |
| Gene (human) | HMG20A (GeneCards) |
| Gene synonyms (human) | HMGX1, HMGXB1 |
| Gene (mouse) | Hmg20a |
| Gene synonyms (mouse) | Ibraf |
| LOGO |  |
| LOGO (reverse complement) | 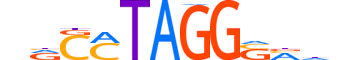 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 12 |
| Consensus | nhbSCCTAKGYn |
| GC content | 55.08% |
| Information content (bits; total / per base) | 11.506 / 0.959 |
| Data sources | HT-SELEX + PBM |
| Aligned words | 714 |
| Previous names |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 1 experiments | median | 0.826 | 0.767 | 0.757 | 0.707 | 0.681 | 0.652 |
| best | 0.826 | 0.767 | 0.757 | 0.707 | 0.681 | 0.652 | |
| PBM benchmarking | auROC, QNZS | auPR, QNZS | auROC, SD | auPR, SD | |
|---|---|---|---|---|---|
| Overall, 8 experiments | median | 0.603 | 0.004 | 0.513 | 0.004 |
| best | 0.737 | 0.008 | 0.595 | 0.017 | |
| TF superclass | Other all-alpha-helical DNA-binding domains {4} (TFClass) |
| TF class | High-mobility group (HMG) domain factors {4.1} (TFClass) |
| TF family | TOX-related {4.1.2} (TFClass) |
| TF subfamily | {4.1.2.0} (TFClass) |
| TFClass ID | TFClass: 4.1.2.0.6 |
| HGNC | HGNC:5001 |
| MGI | MGI:1914117 |
| EntrezGene (human) | GeneID:10363 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:66867 (SSTAR profile) |
| UniProt ID (human) | HM20A_HUMAN |
| UniProt ID (mouse) | HM20A_MOUSE |
| UniProt AC (human) | Q9NP66 (TFClass) |
| UniProt AC (mouse) | Q9DC33 (TFClass) |
| GRECO-DB-TF | no |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 overall: 0 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 8 |
| PCM | HMG20A.H13CORE.0.SB.A.pcm |
| PWM | HMG20A.H13CORE.0.SB.A.pwm |
| PFM | HMG20A.H13CORE.0.SB.A.pfm |
| Threshold to P-value map | HMG20A.H13CORE.0.SB.A.thr |
| Motif in other formats | |
| JASPAR format | HMG20A.H13CORE.0.SB.A_jaspar_format.txt |
| MEME format | HMG20A.H13CORE.0.SB.A_meme_format.meme |
| Transfac format | HMG20A.H13CORE.0.SB.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 161.5 | 178.5 | 135.5 | 238.5 |
| 02 | 147.0 | 120.0 | 114.0 | 333.0 |
| 03 | 62.0 | 100.0 | 138.0 | 414.0 |
| 04 | 2.0 | 419.0 | 186.0 | 107.0 |
| 05 | 0.0 | 714.0 | 0.0 | 0.0 |
| 06 | 0.0 | 699.0 | 0.0 | 15.0 |
| 07 | 0.0 | 0.0 | 0.0 | 714.0 |
| 08 | 714.0 | 0.0 | 0.0 | 0.0 |
| 09 | 3.0 | 3.0 | 427.0 | 281.0 |
| 10 | 34.0 | 115.0 | 562.0 | 3.0 |
| 11 | 61.0 | 418.0 | 74.0 | 161.0 |
| 12 | 224.0 | 183.0 | 133.0 | 174.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.226 | 0.25 | 0.19 | 0.334 |
| 02 | 0.206 | 0.168 | 0.16 | 0.466 |
| 03 | 0.087 | 0.14 | 0.193 | 0.58 |
| 04 | 0.003 | 0.587 | 0.261 | 0.15 |
| 05 | 0.0 | 1.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.979 | 0.0 | 0.021 |
| 07 | 0.0 | 0.0 | 0.0 | 1.0 |
| 08 | 1.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.004 | 0.004 | 0.598 | 0.394 |
| 10 | 0.048 | 0.161 | 0.787 | 0.004 |
| 11 | 0.085 | 0.585 | 0.104 | 0.225 |
| 12 | 0.314 | 0.256 | 0.186 | 0.244 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.099 | 0.0 | -0.273 | 0.287 |
| 02 | -0.192 | -0.393 | -0.443 | 0.619 |
| 03 | -1.04 | -0.572 | -0.255 | 0.836 |
| 04 | -3.901 | 0.848 | 0.041 | -0.506 |
| 05 | -4.697 | 1.379 | -4.697 | -4.697 |
| 06 | -4.697 | 1.358 | -4.697 | -2.382 |
| 07 | -4.697 | -4.697 | -4.697 | 1.379 |
| 08 | 1.379 | -4.697 | -4.697 | -4.697 |
| 09 | -3.658 | -3.658 | 0.867 | 0.45 |
| 10 | -1.62 | -0.435 | 1.141 | -3.658 |
| 11 | -1.056 | 0.846 | -0.868 | -0.102 |
| 12 | 0.225 | 0.025 | -0.291 | -0.025 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.08141 |
| 0.0005 | 5.512645 |
| 0.0001 | 7.65894 |