| Motif | MSX2.H13RSNP.1.SM.B |
| Gene (human) | MSX2 (GeneCards) |
| Gene synonyms (human) | HOX8 |
| Gene (mouse) | Msx2 |
| Gene synonyms (mouse) | Hox-8.1, Msx-2 |
| LOGO |  |
| LOGO (reverse complement) | 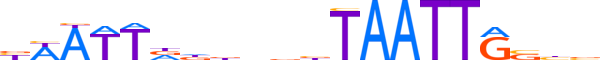 |
| Motif subtype | 1 |
| Quality | B |
| Motif | MSX2.H13RSNP.1.SM.B |
| Gene (human) | MSX2 (GeneCards) |
| Gene synonyms (human) | HOX8 |
| Gene (mouse) | Msx2 |
| Gene synonyms (mouse) | Hox-8.1, Msx-2 |
| LOGO |  |
| LOGO (reverse complement) | 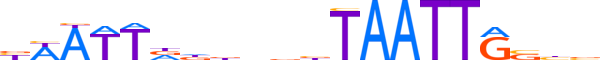 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 20 |
| Consensus | vvbCAATTAvvnhhhAATWv |
| GC content | 31.96% |
| Information content (bits; total / per base) | 17.159 / 0.858 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 252 |
| Previous names | MSX2.H12RSNP.1.SM.B |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.851 | 0.849 | 0.783 | 0.784 | 0.743 | 0.742 |
| best | 0.987 | 0.98 | 0.977 | 0.965 | 0.96 | 0.942 | |
| Methyl HT-SELEX, 1 experiments | median | 0.982 | 0.972 | 0.968 | 0.951 | 0.941 | 0.919 |
| best | 0.982 | 0.972 | 0.968 | 0.951 | 0.941 | 0.919 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.721 | 0.725 | 0.598 | 0.616 | 0.544 | 0.565 |
| best | 0.987 | 0.98 | 0.977 | 0.965 | 0.96 | 0.942 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.601 | 0.046 | 0.556 | 0.373 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | MSX {3.1.2.11} (TFClass) |
| TFClass ID | TFClass: 3.1.2.11.2 |
| HGNC | HGNC:7392 |
| MGI | MGI:97169 |
| EntrezGene (human) | GeneID:4488 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17702 (SSTAR profile) |
| UniProt ID (human) | MSX2_HUMAN |
| UniProt ID (mouse) | MSX2_MOUSE |
| UniProt AC (human) | P35548 (TFClass) |
| UniProt AC (mouse) | Q03358 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | MSX2.H13RSNP.1.SM.B.pcm |
| PWM | MSX2.H13RSNP.1.SM.B.pwm |
| PFM | MSX2.H13RSNP.1.SM.B.pfm |
| Threshold to P-value map | MSX2.H13RSNP.1.SM.B.thr |
| Motif in other formats | |
| JASPAR format | MSX2.H13RSNP.1.SM.B_jaspar_format.txt |
| MEME format | MSX2.H13RSNP.1.SM.B_meme_format.meme |
| Transfac format | MSX2.H13RSNP.1.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 39.25 | 65.25 | 110.25 | 37.25 |
| 02 | 64.5 | 41.5 | 116.5 | 29.5 |
| 03 | 19.0 | 122.0 | 91.0 | 20.0 |
| 04 | 0.0 | 176.0 | 1.0 | 75.0 |
| 05 | 252.0 | 0.0 | 0.0 | 0.0 |
| 06 | 252.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 252.0 |
| 08 | 0.0 | 0.0 | 0.0 | 252.0 |
| 09 | 239.0 | 0.0 | 13.0 | 0.0 |
| 10 | 102.0 | 63.0 | 62.0 | 25.0 |
| 11 | 62.0 | 105.0 | 43.0 | 42.0 |
| 12 | 76.0 | 82.0 | 32.0 | 62.0 |
| 13 | 112.0 | 62.0 | 37.0 | 41.0 |
| 14 | 71.0 | 115.0 | 25.0 | 41.0 |
| 15 | 59.0 | 92.0 | 2.0 | 99.0 |
| 16 | 208.0 | 4.0 | 0.0 | 40.0 |
| 17 | 191.0 | 1.0 | 2.0 | 58.0 |
| 18 | 18.0 | 8.0 | 8.0 | 218.0 |
| 19 | 57.25 | 8.25 | 22.25 | 164.25 |
| 20 | 132.0 | 48.0 | 53.0 | 19.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.156 | 0.259 | 0.438 | 0.148 |
| 02 | 0.256 | 0.165 | 0.462 | 0.117 |
| 03 | 0.075 | 0.484 | 0.361 | 0.079 |
| 04 | 0.0 | 0.698 | 0.004 | 0.298 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 1.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 1.0 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 |
| 09 | 0.948 | 0.0 | 0.052 | 0.0 |
| 10 | 0.405 | 0.25 | 0.246 | 0.099 |
| 11 | 0.246 | 0.417 | 0.171 | 0.167 |
| 12 | 0.302 | 0.325 | 0.127 | 0.246 |
| 13 | 0.444 | 0.246 | 0.147 | 0.163 |
| 14 | 0.282 | 0.456 | 0.099 | 0.163 |
| 15 | 0.234 | 0.365 | 0.008 | 0.393 |
| 16 | 0.825 | 0.016 | 0.0 | 0.159 |
| 17 | 0.758 | 0.004 | 0.008 | 0.23 |
| 18 | 0.071 | 0.032 | 0.032 | 0.865 |
| 19 | 0.227 | 0.033 | 0.088 | 0.652 |
| 20 | 0.524 | 0.19 | 0.21 | 0.075 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.46 | 0.034 | 0.55 | -0.511 |
| 02 | 0.023 | -0.406 | 0.605 | -0.735 |
| 03 | -1.15 | 0.65 | 0.361 | -1.102 |
| 04 | -3.841 | 1.013 | -3.297 | 0.171 |
| 05 | 1.37 | -3.841 | -3.841 | -3.841 |
| 06 | 1.37 | -3.841 | -3.841 | -3.841 |
| 07 | -3.841 | -3.841 | -3.841 | 1.37 |
| 08 | -3.841 | -3.841 | -3.841 | 1.37 |
| 09 | 1.317 | -3.841 | -1.499 | -3.841 |
| 10 | 0.474 | 0.0 | -0.016 | -0.892 |
| 11 | -0.016 | 0.502 | -0.372 | -0.395 |
| 12 | 0.184 | 0.259 | -0.657 | -0.016 |
| 13 | 0.566 | -0.016 | -0.517 | -0.418 |
| 14 | 0.117 | 0.592 | -0.892 | -0.418 |
| 15 | -0.064 | 0.372 | -2.946 | 0.444 |
| 16 | 1.179 | -2.482 | -3.841 | -0.442 |
| 17 | 1.095 | -3.297 | -2.946 | -0.081 |
| 18 | -1.2 | -1.926 | -1.926 | 1.226 |
| 19 | -0.094 | -1.9 | -1.002 | 0.945 |
| 20 | 0.728 | -0.265 | -0.169 | -1.15 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.94451 |
| 0.0005 | 3.27826 |
| 0.0001 | 6.06831 |