| Motif | PRDM5.H13RSNP.0.PSGI.D |
| Gene (human) | PRDM5 (GeneCards) |
| Gene synonyms (human) | PFM2 |
| Gene (mouse) | Prdm5 |
| Gene synonyms (mouse) | |
| LOGO | 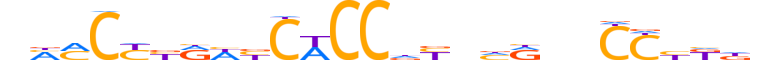 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | PRDM5.H13RSNP.0.PSGI.D |
| Gene (human) | PRDM5 (GeneCards) |
| Gene synonyms (human) | PFM2 |
| Gene (mouse) | Prdm5 |
| Gene synonyms (mouse) | |
| LOGO | 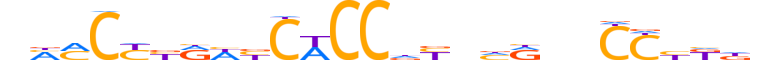 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 26 |
| Consensus | nMMCYbRWbCWCCdYnhRnnCChKdn |
| GC content | 58.24% |
| Information content (bits; total / per base) | 16.433 / 0.632 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX + SMiLe-Seq |
| Aligned words | 2325 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (3) | 0.816 | 0.851 | 0.691 | 0.762 | 0.723 | 0.749 | 97.201 | 136.509 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| overall, 4 experiments | median | 0.997 | 0.996 | 0.958 | 0.956 | 0.863 | 0.878 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.995 | 0.993 | |
| Lysate, 2 experiments | median | 0.99 | 0.986 | 0.89 | 0.889 | 0.716 | 0.746 |
| best | 0.995 | 0.993 | 0.917 | 0.915 | 0.733 | 0.765 | |
| GFPIVT, 2 experiments | median | 1.0 | 0.999 | 0.999 | 0.998 | 0.994 | 0.992 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.995 | 0.993 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| overall, 4 experiments | median | 335.825 | 0.886 | 0.878 | 0.778 |
| best | 516.409 | 0.903 | 0.889 | 0.82 | |
| Lysate, 2 experiments | median | 319.878 | 0.851 | 0.834 | 0.733 |
| best | 392.796 | 0.884 | 0.881 | 0.78 | |
| GFPIVT, 2 experiments | median | 353.484 | 0.893 | 0.878 | 0.79 |
| best | 516.409 | 0.903 | 0.889 | 0.82 | |
| SMiLE-Seq benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.852 | 0.843 | 0.724 | 0.736 | 0.628 | 0.651 |
| best | 0.97 | 0.958 | 0.888 | 0.872 | 0.745 | 0.752 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.195 |
| HGNC | HGNC:9349 |
| MGI | MGI:1918029 |
| EntrezGene (human) | GeneID:11107 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:70779 (SSTAR profile) |
| UniProt ID (human) | PRDM5_HUMAN |
| UniProt ID (mouse) | PRDM5_MOUSE |
| UniProt AC (human) | Q9NQX1 (TFClass) |
| UniProt AC (mouse) | Q9CXE0 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 4 overall: 2 Lysate, 0 IVT, 2 GFPIVT |
| Genomic HT-SELEX | 4 overall: 2 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 2 |
| PBM | 0 |
| PCM | PRDM5.H13RSNP.0.PSGI.D.pcm |
| PWM | PRDM5.H13RSNP.0.PSGI.D.pwm |
| PFM | PRDM5.H13RSNP.0.PSGI.D.pfm |
| Threshold to P-value map | PRDM5.H13RSNP.0.PSGI.D.thr |
| Motif in other formats | |
| JASPAR format | PRDM5.H13RSNP.0.PSGI.D_jaspar_format.txt |
| MEME format | PRDM5.H13RSNP.0.PSGI.D_meme_format.meme |
| Transfac format | PRDM5.H13RSNP.0.PSGI.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 682.75 | 571.75 | 489.75 | 580.75 |
| 02 | 1441.0 | 331.0 | 259.0 | 294.0 |
| 03 | 1003.0 | 1156.0 | 102.0 | 64.0 |
| 04 | 53.0 | 2224.0 | 16.0 | 32.0 |
| 05 | 188.0 | 1327.0 | 55.0 | 755.0 |
| 06 | 112.0 | 498.0 | 474.0 | 1241.0 |
| 07 | 535.0 | 146.0 | 1454.0 | 190.0 |
| 08 | 1382.0 | 389.0 | 109.0 | 445.0 |
| 09 | 68.0 | 768.0 | 412.0 | 1077.0 |
| 10 | 68.0 | 2126.0 | 8.0 | 123.0 |
| 11 | 1205.0 | 25.0 | 43.0 | 1052.0 |
| 12 | 1.0 | 2324.0 | 0.0 | 0.0 |
| 13 | 0.0 | 2325.0 | 0.0 | 0.0 |
| 14 | 718.0 | 175.0 | 933.0 | 499.0 |
| 15 | 52.0 | 638.0 | 353.0 | 1282.0 |
| 16 | 287.0 | 686.0 | 584.0 | 768.0 |
| 17 | 462.0 | 1267.0 | 271.0 | 325.0 |
| 18 | 447.0 | 173.0 | 1466.0 | 239.0 |
| 19 | 721.0 | 564.0 | 645.0 | 395.0 |
| 20 | 699.0 | 588.0 | 632.0 | 406.0 |
| 21 | 100.0 | 2045.0 | 77.0 | 103.0 |
| 22 | 138.0 | 1892.0 | 127.0 | 168.0 |
| 23 | 192.0 | 879.0 | 189.0 | 1065.0 |
| 24 | 233.0 | 178.0 | 540.0 | 1374.0 |
| 25 | 261.5 | 232.5 | 1161.5 | 669.5 |
| 26 | 618.0 | 474.0 | 802.0 | 431.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.294 | 0.246 | 0.211 | 0.25 |
| 02 | 0.62 | 0.142 | 0.111 | 0.126 |
| 03 | 0.431 | 0.497 | 0.044 | 0.028 |
| 04 | 0.023 | 0.957 | 0.007 | 0.014 |
| 05 | 0.081 | 0.571 | 0.024 | 0.325 |
| 06 | 0.048 | 0.214 | 0.204 | 0.534 |
| 07 | 0.23 | 0.063 | 0.625 | 0.082 |
| 08 | 0.594 | 0.167 | 0.047 | 0.191 |
| 09 | 0.029 | 0.33 | 0.177 | 0.463 |
| 10 | 0.029 | 0.914 | 0.003 | 0.053 |
| 11 | 0.518 | 0.011 | 0.018 | 0.452 |
| 12 | 0.0 | 1.0 | 0.0 | 0.0 |
| 13 | 0.0 | 1.0 | 0.0 | 0.0 |
| 14 | 0.309 | 0.075 | 0.401 | 0.215 |
| 15 | 0.022 | 0.274 | 0.152 | 0.551 |
| 16 | 0.123 | 0.295 | 0.251 | 0.33 |
| 17 | 0.199 | 0.545 | 0.117 | 0.14 |
| 18 | 0.192 | 0.074 | 0.631 | 0.103 |
| 19 | 0.31 | 0.243 | 0.277 | 0.17 |
| 20 | 0.301 | 0.253 | 0.272 | 0.175 |
| 21 | 0.043 | 0.88 | 0.033 | 0.044 |
| 22 | 0.059 | 0.814 | 0.055 | 0.072 |
| 23 | 0.083 | 0.378 | 0.081 | 0.458 |
| 24 | 0.1 | 0.077 | 0.232 | 0.591 |
| 25 | 0.112 | 0.1 | 0.5 | 0.288 |
| 26 | 0.266 | 0.204 | 0.345 | 0.185 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.16 | -0.016 | -0.171 | -0.001 |
| 02 | 0.906 | -0.561 | -0.804 | -0.678 |
| 03 | 0.544 | 0.686 | -1.725 | -2.18 |
| 04 | -2.362 | 1.339 | -3.482 | -2.844 |
| 05 | -1.122 | 0.824 | -2.327 | 0.261 |
| 06 | -1.633 | -0.154 | -0.203 | 0.757 |
| 07 | -0.083 | -1.372 | 0.915 | -1.111 |
| 08 | 0.864 | -0.4 | -1.66 | -0.266 |
| 09 | -2.121 | 0.278 | -0.343 | 0.615 |
| 10 | -2.121 | 1.294 | -4.072 | -1.541 |
| 11 | 0.727 | -3.075 | -2.563 | 0.592 |
| 12 | -5.291 | 1.383 | -5.707 | -5.707 |
| 13 | -5.707 | 1.384 | -5.707 | -5.707 |
| 14 | 0.211 | -1.193 | 0.472 | -0.152 |
| 15 | -2.381 | 0.093 | -0.497 | 0.789 |
| 16 | -0.702 | 0.165 | 0.005 | 0.278 |
| 17 | -0.229 | 0.777 | -0.759 | -0.579 |
| 18 | -0.262 | -1.204 | 0.923 | -0.884 |
| 19 | 0.215 | -0.03 | 0.104 | -0.385 |
| 20 | 0.184 | 0.012 | 0.083 | -0.357 |
| 21 | -1.744 | 1.256 | -2.0 | -1.715 |
| 22 | -1.427 | 1.178 | -1.509 | -1.233 |
| 23 | -1.101 | 0.412 | -1.117 | 0.604 |
| 24 | -0.909 | -1.176 | -0.073 | 0.858 |
| 25 | -0.795 | -0.911 | 0.691 | 0.141 |
| 26 | 0.061 | -0.203 | 0.321 | -0.298 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.31801 |
| 0.0005 | 3.59126 |
| 0.0001 | 6.20596 |