| Motif | TF65.H13RSNP.0.P.B |
| Gene (human) | RELA (GeneCards) |
| Gene synonyms (human) | NFKB3 |
| Gene (mouse) | Rela |
| Gene synonyms (mouse) | Nfkb3 |
| LOGO | 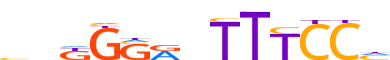 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | TF65.H13RSNP.0.P.B |
| Gene (human) | RELA (GeneCards) |
| Gene synonyms (human) | NFKB3 |
| Gene (mouse) | Rela |
| Gene synonyms (mouse) | Nfkb3 |
| LOGO | 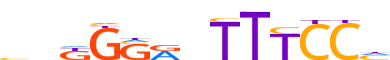 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 13 |
| Consensus | bhKGSRnTTTCCh |
| GC content | 56.22% |
| Information content (bits; total / per base) | 10.538 / 0.811 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | TF65.H12RSNP.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 65 (404) | 0.893 | 0.967 | 0.835 | 0.938 | 0.856 | 0.947 | 3.51 | 4.668 | 229.281 | 576.222 |
| Mouse | 77 (478) | 0.853 | 0.92 | 0.772 | 0.888 | 0.777 | 0.877 | 2.686 | 3.952 | 145.998 | 494.585 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 2.675 | 9.768 | 0.171 | 0.123 |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Rel homology region (RHR) factors {6.1} (TFClass) |
| TF family | NFkappaB-related {6.1.1} (TFClass) |
| TF subfamily | NFkappaB p65-like {6.1.1.2} (TFClass) |
| TFClass ID | TFClass: 6.1.1.2.1 |
| HGNC | HGNC:9955 |
| MGI | MGI:103290 |
| EntrezGene (human) | GeneID:5970 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:19697 (SSTAR profile) |
| UniProt ID (human) | TF65_HUMAN |
| UniProt ID (mouse) | TF65_MOUSE |
| UniProt AC (human) | Q04206 (TFClass) |
| UniProt AC (mouse) | Q04207 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 65 human, 77 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | TF65.H13RSNP.0.P.B.pcm |
| PWM | TF65.H13RSNP.0.P.B.pwm |
| PFM | TF65.H13RSNP.0.P.B.pfm |
| Threshold to P-value map | TF65.H13RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | TF65.H13RSNP.0.P.B_jaspar_format.txt |
| MEME format | TF65.H13RSNP.0.P.B_meme_format.meme |
| Transfac format | TF65.H13RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 147.0 | 453.0 | 208.0 | 192.0 |
| 02 | 219.0 | 385.0 | 146.0 | 250.0 |
| 03 | 78.0 | 91.0 | 618.0 | 213.0 |
| 04 | 36.0 | 44.0 | 865.0 | 55.0 |
| 05 | 95.0 | 100.0 | 731.0 | 74.0 |
| 06 | 520.0 | 41.0 | 383.0 | 56.0 |
| 07 | 333.0 | 295.0 | 180.0 | 192.0 |
| 08 | 13.0 | 87.0 | 6.0 | 894.0 |
| 09 | 2.0 | 25.0 | 18.0 | 955.0 |
| 10 | 4.0 | 205.0 | 17.0 | 774.0 |
| 11 | 12.0 | 878.0 | 16.0 | 94.0 |
| 12 | 6.0 | 891.0 | 22.0 | 81.0 |
| 13 | 260.0 | 514.0 | 90.0 | 136.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.147 | 0.453 | 0.208 | 0.192 |
| 02 | 0.219 | 0.385 | 0.146 | 0.25 |
| 03 | 0.078 | 0.091 | 0.618 | 0.213 |
| 04 | 0.036 | 0.044 | 0.865 | 0.055 |
| 05 | 0.095 | 0.1 | 0.731 | 0.074 |
| 06 | 0.52 | 0.041 | 0.383 | 0.056 |
| 07 | 0.333 | 0.295 | 0.18 | 0.192 |
| 08 | 0.013 | 0.087 | 0.006 | 0.894 |
| 09 | 0.002 | 0.025 | 0.018 | 0.955 |
| 10 | 0.004 | 0.205 | 0.017 | 0.774 |
| 11 | 0.012 | 0.878 | 0.016 | 0.094 |
| 12 | 0.006 | 0.891 | 0.022 | 0.081 |
| 13 | 0.26 | 0.514 | 0.09 | 0.136 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.526 | 0.591 | -0.183 | -0.262 |
| 02 | -0.131 | 0.429 | -0.533 | 0.0 |
| 03 | -1.15 | -0.999 | 0.901 | -0.159 |
| 04 | -1.898 | -1.706 | 1.236 | -1.49 |
| 05 | -0.956 | -0.906 | 1.068 | -1.201 |
| 06 | 0.729 | -1.774 | 0.424 | -1.473 |
| 07 | 0.285 | 0.164 | -0.326 | -0.262 |
| 08 | -2.839 | -1.043 | -3.484 | 1.269 |
| 09 | -4.213 | -2.243 | -2.546 | 1.335 |
| 10 | -3.783 | -0.197 | -2.598 | 1.125 |
| 11 | -2.909 | 1.251 | -2.653 | -0.967 |
| 12 | -3.484 | 1.266 | -2.362 | -1.113 |
| 13 | 0.039 | 0.717 | -1.01 | -0.603 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.60106 |
| 0.0005 | 5.46871 |
| 0.0001 | 7.246595 |