| Motif | THB.H13CORE.1.PSM.A |
| Gene (human) | THRB (GeneCards) |
| Gene synonyms (human) | ERBA2, NR1A2, THR1 |
| Gene (mouse) | Thrb |
| Gene synonyms (mouse) | Erba2, Nr1a2 |
| LOGO |  |
| LOGO (reverse complement) | 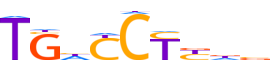 |
| Motif subtype | 1 |
| Quality | A |
| Motif | THB.H13CORE.1.PSM.A |
| Gene (human) | THRB (GeneCards) |
| Gene synonyms (human) | ERBA2, NR1A2, THR1 |
| Gene (mouse) | Thrb |
| Gene synonyms (mouse) | Erba2, Nr1a2 |
| LOGO |  |
| LOGO (reverse complement) | 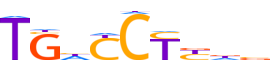 |
| Motif subtype | 1 |
| Quality | A |
| Motif length | 9 |
| Consensus | vhvRGRdCA |
| GC content | 53.9% |
| Information content (bits; total / per base) | 7.13 / 0.792 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 996 |
| Previous names | THB.H12CORE.1.PSM.A |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.795 | 0.809 | 0.622 | 0.642 | 0.793 | 0.795 | 2.294 | 2.311 | 134.051 | 154.585 |
| Mouse | 1 (5) | 0.643 | 0.677 | 0.494 | 0.531 | 0.599 | 0.64 | 1.478 | 1.64 | 17.585 | 33.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.985 | 0.973 | 0.976 | 0.962 | 0.947 | 0.929 |
| best | 0.991 | 0.984 | 0.987 | 0.977 | 0.972 | 0.959 | |
| Methyl HT-SELEX, 2 experiments | median | 0.985 | 0.975 | 0.976 | 0.962 | 0.947 | 0.929 |
| best | 0.991 | 0.984 | 0.985 | 0.976 | 0.964 | 0.951 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.844 | 0.833 | 0.787 | 0.787 | 0.757 | 0.757 |
| best | 0.99 | 0.983 | 0.987 | 0.977 | 0.972 | 0.959 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Thyroid hormone receptor-related {2.1.2} (TFClass) |
| TF subfamily | T3R (NR1A) {2.1.2.2} (TFClass) |
| TFClass ID | TFClass: 2.1.2.2.2 |
| HGNC | HGNC:11799 |
| MGI | MGI:98743 |
| EntrezGene (human) | GeneID:7068 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21834 (SSTAR profile) |
| UniProt ID (human) | THB_HUMAN |
| UniProt ID (mouse) | THB_MOUSE |
| UniProt AC (human) | P10828 (TFClass) |
| UniProt AC (mouse) | P37242 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 1 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | THB.H13CORE.1.PSM.A.pcm |
| PWM | THB.H13CORE.1.PSM.A.pwm |
| PFM | THB.H13CORE.1.PSM.A.pfm |
| Threshold to P-value map | THB.H13CORE.1.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | THB.H13CORE.1.PSM.A_jaspar_format.txt |
| MEME format | THB.H13CORE.1.PSM.A_meme_format.meme |
| Transfac format | THB.H13CORE.1.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 164.0 | 458.0 | 247.0 | 127.0 |
| 02 | 281.0 | 214.0 | 82.0 | 419.0 |
| 03 | 299.0 | 182.0 | 492.0 | 23.0 |
| 04 | 714.0 | 1.0 | 242.0 | 39.0 |
| 05 | 22.0 | 1.0 | 939.0 | 34.0 |
| 06 | 106.0 | 29.0 | 782.0 | 79.0 |
| 07 | 256.0 | 106.0 | 131.0 | 503.0 |
| 08 | 59.0 | 820.0 | 73.0 | 44.0 |
| 09 | 921.0 | 12.0 | 21.0 | 42.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.165 | 0.46 | 0.248 | 0.128 |
| 02 | 0.282 | 0.215 | 0.082 | 0.421 |
| 03 | 0.3 | 0.183 | 0.494 | 0.023 |
| 04 | 0.717 | 0.001 | 0.243 | 0.039 |
| 05 | 0.022 | 0.001 | 0.943 | 0.034 |
| 06 | 0.106 | 0.029 | 0.785 | 0.079 |
| 07 | 0.257 | 0.106 | 0.132 | 0.505 |
| 08 | 0.059 | 0.823 | 0.073 | 0.044 |
| 09 | 0.925 | 0.012 | 0.021 | 0.042 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.414 | 0.606 | -0.008 | -0.667 |
| 02 | 0.12 | -0.15 | -1.097 | 0.518 |
| 03 | 0.182 | -0.311 | 0.678 | -2.317 |
| 04 | 1.049 | -4.522 | -0.028 | -1.817 |
| 05 | -2.358 | -4.522 | 1.322 | -1.948 |
| 06 | -0.845 | -2.099 | 1.14 | -1.133 |
| 07 | 0.028 | -0.845 | -0.636 | 0.7 |
| 08 | -1.418 | 1.187 | -1.211 | -1.702 |
| 09 | 1.303 | -2.905 | -2.401 | -1.746 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.097415 |
| 0.0005 | 5.771545 |
| 0.0001 | 6.934685 |