| Motif | TIGD3.H13CORE.0.PSGIB.A |
| Gene (human) | TIGD3 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tigd3 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 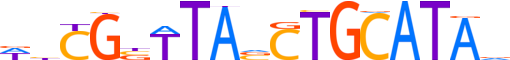 |
| Motif subtype | 0 |
| Quality | A |
| Motif | TIGD3.H13CORE.0.PSGIB.A |
| Gene (human) | TIGD3 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tigd3 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 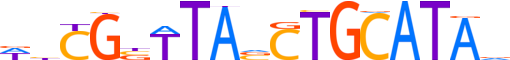 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 17 |
| Consensus | bTATGCAGbTAAvCGdW |
| GC content | 42.84% |
| Information content (bits; total / per base) | 21.336 / 1.255 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX + SMiLe-Seq + PBM |
| Aligned words | 8794 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 3 (3) | 0.838 | 0.851 | 0.741 | 0.762 | 0.686 | 0.722 | 81.347 | 99.602 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| overall, 3 experiments | median | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Lysate, 2 experiments | median | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| GFPIVT, 1 experiments | median | 1.0 | 1.0 | 0.967 | 0.969 | 0.752 | 0.798 |
| best | 1.0 | 1.0 | 0.967 | 0.969 | 0.752 | 0.798 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| Lysate, 2 experiments | median | 537.638 | 0.971 | 0.973 | 0.965 |
| best | 750.824 | 0.976 | 0.983 | 0.972 | |
| SMiLE-Seq benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 1 experiments | median | 0.827 | 0.809 | 0.682 | 0.683 | 0.594 | 0.61 |
| best | 0.827 | 0.809 | 0.682 | 0.683 | 0.594 | 0.61 | |
| PBM benchmarking | auROC, QNZS | auPR, QNZS | auROC, SD | auPR, SD | |
|---|---|---|---|---|---|
| Overall, 10 experiments | median | 0.747 | 0.034 | 0.679 | 0.051 |
| best | 0.788 | 0.054 | 0.811 | 0.059 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Other {3.0} (TFClass) |
| TF family | CENPB {3.0.256} (TFClass) |
| TF subfamily | {3.0.256.0} (TFClass) |
| TFClass ID | TFClass: 3.0.256.0.2 |
| HGNC | |
| MGI | |
| EntrezGene (human) | |
| EntrezGene (mouse) | |
| UniProt ID (human) | TIGD3_HUMAN |
| UniProt ID (mouse) | TIGD3_MOUSE |
| UniProt AC (human) | Q6B0B8 (TFClass) |
| UniProt AC (mouse) | Q7TM95 (TFClass) |
| GRECO-DB-TF | no |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 3 overall: 2 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 2 overall: 2 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 1 |
| PBM | 10 |
| PCM | TIGD3.H13CORE.0.PSGIB.A.pcm |
| PWM | TIGD3.H13CORE.0.PSGIB.A.pwm |
| PFM | TIGD3.H13CORE.0.PSGIB.A.pfm |
| Threshold to P-value map | TIGD3.H13CORE.0.PSGIB.A.thr |
| Motif in other formats | |
| JASPAR format | TIGD3.H13CORE.0.PSGIB.A_jaspar_format.txt |
| MEME format | TIGD3.H13CORE.0.PSGIB.A_meme_format.meme |
| Transfac format | TIGD3.H13CORE.0.PSGIB.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1271.25 | 2158.25 | 1318.25 | 4046.25 |
| 02 | 159.25 | 324.25 | 172.25 | 8138.25 |
| 03 | 8774.0 | 2.0 | 8.0 | 10.0 |
| 04 | 16.0 | 8.0 | 0.0 | 8770.0 |
| 05 | 30.0 | 185.0 | 8369.0 | 210.0 |
| 06 | 5.0 | 8782.0 | 1.0 | 6.0 |
| 07 | 8387.0 | 27.0 | 265.0 | 115.0 |
| 08 | 12.0 | 1660.0 | 7101.0 | 21.0 |
| 09 | 268.0 | 2702.0 | 3986.0 | 1838.0 |
| 10 | 200.0 | 7.0 | 8.0 | 8579.0 |
| 11 | 8769.0 | 1.0 | 13.0 | 11.0 |
| 12 | 6699.0 | 36.0 | 42.0 | 2017.0 |
| 13 | 2235.0 | 3792.0 | 2721.0 | 46.0 |
| 14 | 220.0 | 8327.0 | 211.0 | 36.0 |
| 15 | 1770.0 | 24.0 | 6976.0 | 24.0 |
| 16 | 3310.0 | 838.0 | 2627.0 | 2019.0 |
| 17 | 4306.75 | 1162.75 | 187.75 | 3136.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.145 | 0.245 | 0.15 | 0.46 |
| 02 | 0.018 | 0.037 | 0.02 | 0.925 |
| 03 | 0.998 | 0.0 | 0.001 | 0.001 |
| 04 | 0.002 | 0.001 | 0.0 | 0.997 |
| 05 | 0.003 | 0.021 | 0.952 | 0.024 |
| 06 | 0.001 | 0.999 | 0.0 | 0.001 |
| 07 | 0.954 | 0.003 | 0.03 | 0.013 |
| 08 | 0.001 | 0.189 | 0.807 | 0.002 |
| 09 | 0.03 | 0.307 | 0.453 | 0.209 |
| 10 | 0.023 | 0.001 | 0.001 | 0.976 |
| 11 | 0.997 | 0.0 | 0.001 | 0.001 |
| 12 | 0.762 | 0.004 | 0.005 | 0.229 |
| 13 | 0.254 | 0.431 | 0.309 | 0.005 |
| 14 | 0.025 | 0.947 | 0.024 | 0.004 |
| 15 | 0.201 | 0.003 | 0.793 | 0.003 |
| 16 | 0.376 | 0.095 | 0.299 | 0.23 |
| 17 | 0.49 | 0.132 | 0.021 | 0.357 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.547 | -0.018 | -0.511 | 0.61 |
| 02 | -2.612 | -1.908 | -2.535 | 1.308 |
| 03 | 1.383 | -6.245 | -5.367 | -5.189 |
| 04 | -4.791 | -5.367 | -6.877 | 1.383 |
| 05 | -4.222 | -2.464 | 1.336 | -2.339 |
| 06 | -5.713 | 1.384 | -6.512 | -5.584 |
| 07 | 1.338 | -4.32 | -2.108 | -2.932 |
| 08 | -5.038 | -0.281 | 1.172 | -4.549 |
| 09 | -2.097 | 0.206 | 0.595 | -0.179 |
| 10 | -2.387 | -5.47 | -5.367 | 1.361 |
| 11 | 1.383 | -6.512 | -4.971 | -5.111 |
| 12 | 1.113 | -4.052 | -3.906 | -0.086 |
| 13 | 0.016 | 0.545 | 0.213 | -3.82 |
| 14 | -2.293 | 1.331 | -2.334 | -4.052 |
| 15 | -0.217 | -4.428 | 1.154 | -4.428 |
| 16 | 0.409 | -0.963 | 0.178 | -0.085 |
| 17 | 0.672 | -0.636 | -2.449 | 0.355 |
| P-value | Threshold |
|---|---|
| 0.001 | -3.45014 |
| 0.0005 | -1.55119 |
| 0.0001 | 2.43451 |