| Motif | ZBT22.H13CORE.0.SM.B |
| Gene (human) | ZBTB22 (GeneCards) |
| Gene synonyms (human) | BING1, ZBTB22A, ZNF297 |
| Gene (mouse) | Zbtb22 |
| Gene synonyms (mouse) | Bing1, Zfp297, Znf297 |
| LOGO | 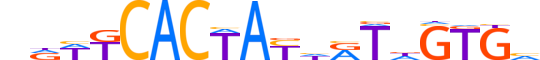 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | ZBT22.H13CORE.0.SM.B |
| Gene (human) | ZBTB22 (GeneCards) |
| Gene synonyms (human) | BING1, ZBTB22A, ZNF297 |
| Gene (mouse) | Zbtb22 |
| Gene synonyms (mouse) | Bing1, Zfp297, Znf297 |
| LOGO | 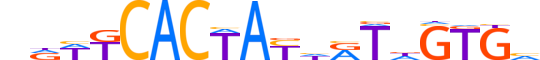 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 18 |
| Consensus | nRWKCACTAYhRTdGTGv |
| GC content | 41.79% |
| Information content (bits; total / per base) | 18.301 / 1.017 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 5235 |
| Previous names | ZBT22.H12CORE.0.SM.B |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.943 | 0.936 | 0.83 | 0.82 | 0.712 | 0.717 |
| best | 0.966 | 0.959 | 0.884 | 0.867 | 0.766 | 0.762 | |
| Methyl HT-SELEX, 1 experiments | median | 0.921 | 0.912 | 0.776 | 0.773 | 0.657 | 0.672 |
| best | 0.921 | 0.912 | 0.776 | 0.773 | 0.657 | 0.672 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.966 | 0.959 | 0.884 | 0.867 | 0.766 | 0.762 |
| best | 0.966 | 0.959 | 0.884 | 0.867 | 0.766 | 0.762 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Other with up to three adjacent zinc fingers {2.3.2} (TFClass) |
| TF subfamily | BTB-POZ {2.3.2.1} (TFClass) |
| TFClass ID | TFClass: 2.3.2.1.6 |
| HGNC | HGNC:13085 |
| MGI | MGI:1931870 |
| EntrezGene (human) | GeneID:9278 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:81630 (SSTAR profile) |
| UniProt ID (human) | ZBT22_HUMAN |
| UniProt ID (mouse) | ZBT22_MOUSE |
| UniProt AC (human) | O15209 (TFClass) |
| UniProt AC (mouse) | Q9Z0G7 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZBT22.H13CORE.0.SM.B.pcm |
| PWM | ZBT22.H13CORE.0.SM.B.pwm |
| PFM | ZBT22.H13CORE.0.SM.B.pfm |
| Threshold to P-value map | ZBT22.H13CORE.0.SM.B.thr |
| Motif in other formats | |
| JASPAR format | ZBT22.H13CORE.0.SM.B_jaspar_format.txt |
| MEME format | ZBT22.H13CORE.0.SM.B_meme_format.meme |
| Transfac format | ZBT22.H13CORE.0.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1124.25 | 1113.25 | 1347.25 | 1650.25 |
| 02 | 1110.0 | 197.0 | 2974.0 | 954.0 |
| 03 | 1012.0 | 787.0 | 80.0 | 3356.0 |
| 04 | 18.0 | 1.0 | 2085.0 | 3131.0 |
| 05 | 0.0 | 5235.0 | 0.0 | 0.0 |
| 06 | 5234.0 | 1.0 | 0.0 | 0.0 |
| 07 | 0.0 | 5235.0 | 0.0 | 0.0 |
| 08 | 575.0 | 287.0 | 91.0 | 4282.0 |
| 09 | 5225.0 | 8.0 | 1.0 | 1.0 |
| 10 | 8.0 | 1387.0 | 536.0 | 3304.0 |
| 11 | 1221.0 | 835.0 | 538.0 | 2641.0 |
| 12 | 2741.0 | 445.0 | 2012.0 | 37.0 |
| 13 | 238.0 | 172.0 | 214.0 | 4611.0 |
| 14 | 2409.0 | 369.0 | 798.0 | 1659.0 |
| 15 | 227.0 | 232.0 | 4596.0 | 180.0 |
| 16 | 169.0 | 131.0 | 458.0 | 4477.0 |
| 17 | 102.75 | 205.75 | 4510.75 | 415.75 |
| 18 | 2286.0 | 1918.0 | 578.0 | 453.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.215 | 0.213 | 0.257 | 0.315 |
| 02 | 0.212 | 0.038 | 0.568 | 0.182 |
| 03 | 0.193 | 0.15 | 0.015 | 0.641 |
| 04 | 0.003 | 0.0 | 0.398 | 0.598 |
| 05 | 0.0 | 1.0 | 0.0 | 0.0 |
| 06 | 1.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 1.0 | 0.0 | 0.0 |
| 08 | 0.11 | 0.055 | 0.017 | 0.818 |
| 09 | 0.998 | 0.002 | 0.0 | 0.0 |
| 10 | 0.002 | 0.265 | 0.102 | 0.631 |
| 11 | 0.233 | 0.16 | 0.103 | 0.504 |
| 12 | 0.524 | 0.085 | 0.384 | 0.007 |
| 13 | 0.045 | 0.033 | 0.041 | 0.881 |
| 14 | 0.46 | 0.07 | 0.152 | 0.317 |
| 15 | 0.043 | 0.044 | 0.878 | 0.034 |
| 16 | 0.032 | 0.025 | 0.087 | 0.855 |
| 17 | 0.02 | 0.039 | 0.862 | 0.079 |
| 18 | 0.437 | 0.366 | 0.11 | 0.087 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.152 | -0.162 | 0.029 | 0.232 |
| 02 | -0.164 | -1.884 | 0.82 | -0.316 |
| 03 | -0.257 | -0.508 | -2.77 | 0.941 |
| 04 | -4.176 | -6.034 | 0.465 | 0.871 |
| 05 | -6.417 | 1.385 | -6.417 | -6.417 |
| 06 | 1.385 | -6.034 | -6.417 | -6.417 |
| 07 | -6.417 | 1.385 | -6.417 | -6.417 |
| 08 | -0.82 | -1.512 | -2.644 | 1.184 |
| 09 | 1.383 | -4.862 | -6.034 | -6.034 |
| 10 | -4.862 | 0.058 | -0.89 | 0.925 |
| 11 | -0.069 | -0.448 | -0.887 | 0.701 |
| 12 | 0.738 | -1.076 | 0.429 | -3.511 |
| 13 | -1.697 | -2.019 | -1.803 | 1.258 |
| 14 | 0.609 | -1.262 | -0.494 | 0.237 |
| 15 | -1.744 | -1.723 | 1.255 | -1.974 |
| 16 | -2.036 | -2.287 | -1.047 | 1.229 |
| 17 | -2.526 | -1.841 | 1.236 | -1.143 |
| 18 | 0.557 | 0.382 | -0.815 | -1.058 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.22619 |
| 0.0005 | 1.49476 |
| 0.0001 | 5.01156 |