| Motif | ZNF234.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF234 (GeneCards) |
| Gene synonyms (human) | ZNF269 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 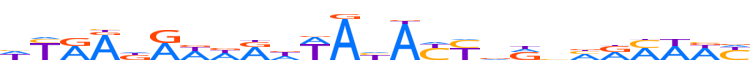 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZNF234.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF234 (GeneCards) |
| Gene synonyms (human) | ZNF269 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 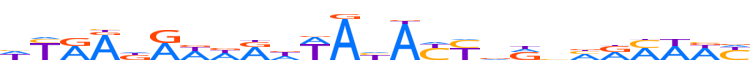 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 25 |
| Consensus | hYRAdAWdRdWAhAYYdRnhRMWWY |
| GC content | 31.81% |
| Information content (bits; total / per base) | 15.504 / 0.62 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX |
| Aligned words | 127 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 3 (3) | 0.789 | 0.809 | 0.767 | 0.77 | 0.875 | 0.879 | 31.658 | 63.0 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 1 experiments | median | 0.474 | 0.488 | 0.485 | 0.491 | 0.492 | 0.494 |
| best | 0.474 | 0.488 | 0.485 | 0.491 | 0.492 | 0.494 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 1 experiments | median | 0.0 | 0.441 | 0.376 | 0.308 |
| best | 1.796 | 0.47 | 0.416 | 0.345 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF222-like {2.3.3.47} (TFClass) |
| TFClass ID | TFClass: 2.3.3.47.14 |
| HGNC | HGNC:13027 |
| MGI | |
| EntrezGene (human) | GeneID:10780 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN234_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q14588 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 1 overall: 0 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 1 overall: 0 Lysate, 0 IVT, 1 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZNF234.H13INVIVO.0.PSG.A.pcm |
| PWM | ZNF234.H13INVIVO.0.PSG.A.pwm |
| PFM | ZNF234.H13INVIVO.0.PSG.A.pfm |
| Threshold to P-value map | ZNF234.H13INVIVO.0.PSG.A.thr |
| Motif in other formats | |
| JASPAR format | ZNF234.H13INVIVO.0.PSG.A_jaspar_format.txt |
| MEME format | ZNF234.H13INVIVO.0.PSG.A_meme_format.meme |
| Transfac format | ZNF234.H13INVIVO.0.PSG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 34.0 | 15.0 | 9.0 | 69.0 |
| 02 | 12.0 | 24.0 | 2.0 | 89.0 |
| 03 | 78.0 | 3.0 | 42.0 | 4.0 |
| 04 | 103.0 | 5.0 | 12.0 | 7.0 |
| 05 | 19.0 | 10.0 | 65.0 | 33.0 |
| 06 | 73.0 | 0.0 | 54.0 | 0.0 |
| 07 | 74.0 | 6.0 | 13.0 | 34.0 |
| 08 | 71.0 | 7.0 | 22.0 | 27.0 |
| 09 | 81.0 | 2.0 | 30.0 | 14.0 |
| 10 | 52.0 | 8.0 | 16.0 | 51.0 |
| 11 | 35.0 | 1.0 | 7.0 | 84.0 |
| 12 | 114.0 | 0.0 | 13.0 | 0.0 |
| 13 | 16.0 | 32.0 | 12.0 | 67.0 |
| 14 | 112.0 | 2.0 | 4.0 | 9.0 |
| 15 | 11.0 | 83.0 | 6.0 | 27.0 |
| 16 | 8.0 | 46.0 | 1.0 | 72.0 |
| 17 | 42.0 | 12.0 | 40.0 | 33.0 |
| 18 | 29.0 | 3.0 | 72.0 | 23.0 |
| 19 | 32.0 | 48.0 | 15.0 | 32.0 |
| 20 | 71.0 | 31.0 | 12.0 | 13.0 |
| 21 | 65.0 | 5.0 | 48.0 | 9.0 |
| 22 | 70.0 | 48.0 | 1.0 | 8.0 |
| 23 | 80.0 | 3.0 | 5.0 | 39.0 |
| 24 | 86.0 | 10.0 | 12.0 | 19.0 |
| 25 | 14.0 | 82.0 | 11.0 | 20.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.268 | 0.118 | 0.071 | 0.543 |
| 02 | 0.094 | 0.189 | 0.016 | 0.701 |
| 03 | 0.614 | 0.024 | 0.331 | 0.031 |
| 04 | 0.811 | 0.039 | 0.094 | 0.055 |
| 05 | 0.15 | 0.079 | 0.512 | 0.26 |
| 06 | 0.575 | 0.0 | 0.425 | 0.0 |
| 07 | 0.583 | 0.047 | 0.102 | 0.268 |
| 08 | 0.559 | 0.055 | 0.173 | 0.213 |
| 09 | 0.638 | 0.016 | 0.236 | 0.11 |
| 10 | 0.409 | 0.063 | 0.126 | 0.402 |
| 11 | 0.276 | 0.008 | 0.055 | 0.661 |
| 12 | 0.898 | 0.0 | 0.102 | 0.0 |
| 13 | 0.126 | 0.252 | 0.094 | 0.528 |
| 14 | 0.882 | 0.016 | 0.031 | 0.071 |
| 15 | 0.087 | 0.654 | 0.047 | 0.213 |
| 16 | 0.063 | 0.362 | 0.008 | 0.567 |
| 17 | 0.331 | 0.094 | 0.315 | 0.26 |
| 18 | 0.228 | 0.024 | 0.567 | 0.181 |
| 19 | 0.252 | 0.378 | 0.118 | 0.252 |
| 20 | 0.559 | 0.244 | 0.094 | 0.102 |
| 21 | 0.512 | 0.039 | 0.378 | 0.071 |
| 22 | 0.551 | 0.378 | 0.008 | 0.063 |
| 23 | 0.63 | 0.024 | 0.039 | 0.307 |
| 24 | 0.677 | 0.079 | 0.094 | 0.15 |
| 25 | 0.11 | 0.646 | 0.087 | 0.157 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.066 | -0.71 | -1.172 | 0.756 |
| 02 | -0.914 | -0.268 | -2.329 | 1.007 |
| 03 | 0.877 | -2.058 | 0.271 | -1.845 |
| 04 | 1.151 | -1.669 | -0.914 | -1.39 |
| 05 | -0.489 | -1.078 | 0.698 | 0.037 |
| 06 | 0.812 | -3.304 | 0.516 | -3.304 |
| 07 | 0.825 | -1.52 | -0.841 | 0.066 |
| 08 | 0.784 | -1.39 | -0.351 | -0.156 |
| 09 | 0.914 | -2.329 | -0.055 | -0.773 |
| 10 | 0.479 | -1.275 | -0.65 | 0.46 |
| 11 | 0.094 | -2.702 | -1.39 | 0.95 |
| 12 | 1.251 | -3.304 | -0.841 | -3.304 |
| 13 | -0.65 | 0.008 | -0.914 | 0.727 |
| 14 | 1.234 | -2.329 | -1.845 | -1.172 |
| 15 | -0.993 | 0.938 | -1.52 | -0.156 |
| 16 | -1.275 | 0.359 | -2.702 | 0.798 |
| 17 | 0.271 | -0.914 | 0.223 | 0.037 |
| 18 | -0.087 | -2.058 | 0.798 | -0.309 |
| 19 | 0.008 | 0.401 | -0.71 | 0.008 |
| 20 | 0.784 | -0.023 | -0.914 | -0.841 |
| 21 | 0.698 | -1.669 | 0.401 | -1.172 |
| 22 | 0.77 | 0.401 | -2.702 | -1.275 |
| 23 | 0.902 | -2.058 | -1.669 | 0.199 |
| 24 | 0.973 | -1.078 | -0.914 | -0.489 |
| 25 | -0.773 | 0.926 | -0.993 | -0.441 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.62051 |
| 0.0005 | 4.62321 |
| 0.0001 | 6.73566 |