| Motif | ZNF510.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF510 (GeneCards) |
| Gene synonyms (human) | KIAA0972 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 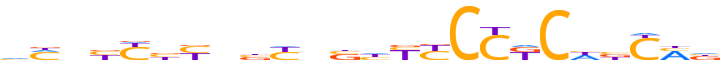 |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZNF510.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF510 (GeneCards) |
| Gene synonyms (human) | KIAA0972 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 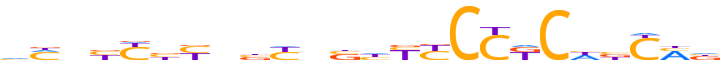 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 24 |
| Consensus | nKRvdGMGGRRdvnRvnRvRvnKb |
| GC content | 58.93% |
| Information content (bits; total / per base) | 12.428 / 0.518 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX |
| Aligned words | 500 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (4) | 0.798 | 0.82 | 0.665 | 0.708 | 0.811 | 0.871 | 41.364 | 50.108 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 0.861 | 0.845 | 0.811 | 0.801 | 0.746 | 0.742 |
| best | 0.997 | 0.995 | 0.985 | 0.981 | 0.911 | 0.908 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 141.614 | 0.837 | 0.795 | 0.697 |
| best | 300.921 | 0.922 | 0.873 | 0.808 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Multiple dispersed zinc fingers {2.3.4} (TFClass) |
| TF subfamily | ZNF37A-like {2.3.4.25} (TFClass) |
| TFClass ID | TFClass: 2.3.4.25.6 |
| HGNC | HGNC:29161 |
| MGI | |
| EntrezGene (human) | GeneID:22869 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN510_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q9Y2H8 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| Genomic HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZNF510.H13INVIVO.0.PSG.A.pcm |
| PWM | ZNF510.H13INVIVO.0.PSG.A.pwm |
| PFM | ZNF510.H13INVIVO.0.PSG.A.pfm |
| Threshold to P-value map | ZNF510.H13INVIVO.0.PSG.A.thr |
| Motif in other formats | |
| JASPAR format | ZNF510.H13INVIVO.0.PSG.A_jaspar_format.txt |
| MEME format | ZNF510.H13INVIVO.0.PSG.A_meme_format.meme |
| Transfac format | ZNF510.H13INVIVO.0.PSG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 82.0 | 267.0 | 82.0 | 69.0 |
| 02 | 61.0 | 24.0 | 120.0 | 295.0 |
| 03 | 51.0 | 17.0 | 391.0 | 41.0 |
| 04 | 188.0 | 178.0 | 120.0 | 14.0 |
| 05 | 156.0 | 30.0 | 64.0 | 250.0 |
| 06 | 3.0 | 8.0 | 482.0 | 7.0 |
| 07 | 265.0 | 171.0 | 11.0 | 53.0 |
| 08 | 115.0 | 3.0 | 377.0 | 5.0 |
| 09 | 2.0 | 7.0 | 490.0 | 1.0 |
| 10 | 181.0 | 39.0 | 266.0 | 14.0 |
| 11 | 318.0 | 69.0 | 86.0 | 27.0 |
| 12 | 181.0 | 34.0 | 200.0 | 85.0 |
| 13 | 60.0 | 277.0 | 126.0 | 37.0 |
| 14 | 98.0 | 148.0 | 106.0 | 148.0 |
| 15 | 167.0 | 27.0 | 268.0 | 38.0 |
| 16 | 94.0 | 210.0 | 160.0 | 36.0 |
| 17 | 127.0 | 63.0 | 153.0 | 157.0 |
| 18 | 228.0 | 32.0 | 221.0 | 19.0 |
| 19 | 209.0 | 74.0 | 179.0 | 38.0 |
| 20 | 109.0 | 26.0 | 334.0 | 31.0 |
| 21 | 241.0 | 52.0 | 172.0 | 35.0 |
| 22 | 87.0 | 84.0 | 171.0 | 158.0 |
| 23 | 61.0 | 54.0 | 306.0 | 79.0 |
| 24 | 56.0 | 101.0 | 191.0 | 152.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.164 | 0.534 | 0.164 | 0.138 |
| 02 | 0.122 | 0.048 | 0.24 | 0.59 |
| 03 | 0.102 | 0.034 | 0.782 | 0.082 |
| 04 | 0.376 | 0.356 | 0.24 | 0.028 |
| 05 | 0.312 | 0.06 | 0.128 | 0.5 |
| 06 | 0.006 | 0.016 | 0.964 | 0.014 |
| 07 | 0.53 | 0.342 | 0.022 | 0.106 |
| 08 | 0.23 | 0.006 | 0.754 | 0.01 |
| 09 | 0.004 | 0.014 | 0.98 | 0.002 |
| 10 | 0.362 | 0.078 | 0.532 | 0.028 |
| 11 | 0.636 | 0.138 | 0.172 | 0.054 |
| 12 | 0.362 | 0.068 | 0.4 | 0.17 |
| 13 | 0.12 | 0.554 | 0.252 | 0.074 |
| 14 | 0.196 | 0.296 | 0.212 | 0.296 |
| 15 | 0.334 | 0.054 | 0.536 | 0.076 |
| 16 | 0.188 | 0.42 | 0.32 | 0.072 |
| 17 | 0.254 | 0.126 | 0.306 | 0.314 |
| 18 | 0.456 | 0.064 | 0.442 | 0.038 |
| 19 | 0.418 | 0.148 | 0.358 | 0.076 |
| 20 | 0.218 | 0.052 | 0.668 | 0.062 |
| 21 | 0.482 | 0.104 | 0.344 | 0.07 |
| 22 | 0.174 | 0.168 | 0.342 | 0.316 |
| 23 | 0.122 | 0.108 | 0.612 | 0.158 |
| 24 | 0.112 | 0.202 | 0.382 | 0.304 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.415 | 0.752 | -0.415 | -0.584 |
| 02 | -0.705 | -1.6 | -0.04 | 0.852 |
| 03 | -0.879 | -1.92 | 1.132 | -1.09 |
| 04 | 0.404 | 0.35 | -0.04 | -2.096 |
| 05 | 0.219 | -1.389 | -0.658 | 0.687 |
| 06 | -3.325 | -2.584 | 1.34 | -2.694 |
| 07 | 0.745 | 0.31 | -2.311 | -0.841 |
| 08 | -0.082 | -3.325 | 1.096 | -2.961 |
| 09 | -3.573 | -2.694 | 1.357 | -3.903 |
| 10 | 0.366 | -1.138 | 0.749 | -2.096 |
| 11 | 0.926 | -0.584 | -0.368 | -1.489 |
| 12 | 0.366 | -1.27 | 0.465 | -0.38 |
| 13 | -0.721 | 0.789 | 0.008 | -1.189 |
| 14 | -0.24 | 0.167 | -0.163 | 0.167 |
| 15 | 0.287 | -1.489 | 0.756 | -1.163 |
| 16 | -0.281 | 0.514 | 0.244 | -1.215 |
| 17 | 0.016 | -0.673 | 0.2 | 0.225 |
| 18 | 0.595 | -1.328 | 0.565 | -1.818 |
| 19 | 0.509 | -0.516 | 0.355 | -1.163 |
| 20 | -0.135 | -1.525 | 0.975 | -1.358 |
| 21 | 0.651 | -0.86 | 0.316 | -1.242 |
| 22 | -0.357 | -0.392 | 0.31 | 0.232 |
| 23 | -0.705 | -0.823 | 0.888 | -0.452 |
| 24 | -0.788 | -0.21 | 0.42 | 0.193 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.19451 |
| 0.0005 | 5.13296 |
| 0.0001 | 7.07406 |