| Motif | ZNF699.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF699 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 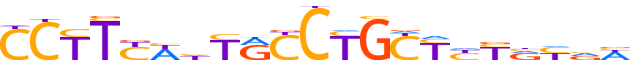 |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZNF699.H13INVIVO.0.PSG.A |
| Gene (human) | ZNF699 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 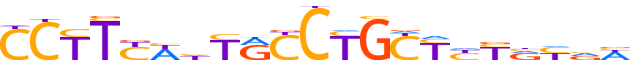 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 21 |
| Consensus | WbRbRRRGCAGGYRdWRAAGG |
| GC content | 53.21% |
| Information content (bits; total / per base) | 17.7 / 0.843 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX |
| Aligned words | 86 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (2) | 0.807 | 0.916 | 0.744 | 0.893 | 0.785 | 0.933 | 17.626 | 24.292 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 1 experiments | median | 0.598 | 0.595 | 0.538 | 0.546 | 0.519 | 0.526 |
| best | 0.598 | 0.595 | 0.538 | 0.546 | 0.519 | 0.526 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 28.72 | 0.678 | 0.557 | 0.535 |
| best | 96.108 | 0.763 | 0.715 | 0.657 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF763-like {2.3.3.33} (TFClass) |
| TFClass ID | TFClass: 2.3.3.33.203 |
| HGNC | HGNC:24750 |
| MGI | |
| EntrezGene (human) | GeneID:374879 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN699_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q32M78 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 1 overall: 0 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZNF699.H13INVIVO.0.PSG.A.pcm |
| PWM | ZNF699.H13INVIVO.0.PSG.A.pwm |
| PFM | ZNF699.H13INVIVO.0.PSG.A.pfm |
| Threshold to P-value map | ZNF699.H13INVIVO.0.PSG.A.thr |
| Motif in other formats | |
| JASPAR format | ZNF699.H13INVIVO.0.PSG.A_jaspar_format.txt |
| MEME format | ZNF699.H13INVIVO.0.PSG.A_meme_format.meme |
| Transfac format | ZNF699.H13INVIVO.0.PSG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 13.0 | 10.0 | 11.0 | 52.0 |
| 02 | 7.0 | 39.0 | 12.0 | 28.0 |
| 03 | 41.0 | 3.0 | 34.0 | 8.0 |
| 04 | 10.0 | 51.0 | 11.0 | 14.0 |
| 05 | 58.0 | 10.0 | 14.0 | 4.0 |
| 06 | 21.0 | 8.0 | 49.0 | 8.0 |
| 07 | 61.0 | 1.0 | 13.0 | 11.0 |
| 08 | 10.0 | 3.0 | 68.0 | 5.0 |
| 09 | 1.0 | 78.0 | 6.0 | 1.0 |
| 10 | 67.0 | 2.0 | 14.0 | 3.0 |
| 11 | 3.0 | 0.0 | 83.0 | 0.0 |
| 12 | 9.0 | 6.0 | 69.0 | 2.0 |
| 13 | 5.0 | 55.0 | 1.0 | 25.0 |
| 14 | 62.0 | 3.0 | 19.0 | 2.0 |
| 15 | 44.0 | 11.0 | 15.0 | 16.0 |
| 16 | 17.0 | 3.0 | 11.0 | 55.0 |
| 17 | 22.0 | 7.0 | 55.0 | 2.0 |
| 18 | 77.0 | 4.0 | 5.0 | 0.0 |
| 19 | 63.0 | 2.0 | 21.0 | 0.0 |
| 20 | 6.0 | 2.0 | 77.0 | 1.0 |
| 21 | 8.0 | 2.0 | 73.0 | 3.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.151 | 0.116 | 0.128 | 0.605 |
| 02 | 0.081 | 0.453 | 0.14 | 0.326 |
| 03 | 0.477 | 0.035 | 0.395 | 0.093 |
| 04 | 0.116 | 0.593 | 0.128 | 0.163 |
| 05 | 0.674 | 0.116 | 0.163 | 0.047 |
| 06 | 0.244 | 0.093 | 0.57 | 0.093 |
| 07 | 0.709 | 0.012 | 0.151 | 0.128 |
| 08 | 0.116 | 0.035 | 0.791 | 0.058 |
| 09 | 0.012 | 0.907 | 0.07 | 0.012 |
| 10 | 0.779 | 0.023 | 0.163 | 0.035 |
| 11 | 0.035 | 0.0 | 0.965 | 0.0 |
| 12 | 0.105 | 0.07 | 0.802 | 0.023 |
| 13 | 0.058 | 0.64 | 0.012 | 0.291 |
| 14 | 0.721 | 0.035 | 0.221 | 0.023 |
| 15 | 0.512 | 0.128 | 0.174 | 0.186 |
| 16 | 0.198 | 0.035 | 0.128 | 0.64 |
| 17 | 0.256 | 0.081 | 0.64 | 0.023 |
| 18 | 0.895 | 0.047 | 0.058 | 0.0 |
| 19 | 0.733 | 0.023 | 0.244 | 0.0 |
| 20 | 0.07 | 0.023 | 0.895 | 0.012 |
| 21 | 0.093 | 0.023 | 0.849 | 0.035 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.471 | -0.71 | -0.624 | 0.854 |
| 02 | -1.025 | 0.573 | -0.545 | 0.253 |
| 03 | 0.622 | -1.704 | 0.44 | -0.909 |
| 04 | -0.71 | 0.835 | -0.624 | -0.403 |
| 05 | 0.961 | -0.71 | -0.403 | -1.487 |
| 06 | -0.022 | -0.909 | 0.796 | -0.909 |
| 07 | 1.01 | -2.37 | -0.471 | -0.624 |
| 08 | -0.71 | -1.704 | 1.117 | -1.308 |
| 09 | -2.37 | 1.252 | -1.157 | -2.37 |
| 10 | 1.103 | -1.983 | -0.403 | -1.704 |
| 11 | -1.704 | -3.011 | 1.314 | -3.011 |
| 12 | -0.805 | -1.157 | 1.132 | -1.983 |
| 13 | -1.308 | 0.909 | -2.37 | 0.144 |
| 14 | 1.026 | -1.704 | -0.117 | -1.983 |
| 15 | 0.691 | -0.624 | -0.339 | -0.279 |
| 16 | -0.222 | -1.704 | -0.624 | 0.909 |
| 17 | 0.022 | -1.025 | 0.909 | -1.983 |
| 18 | 1.24 | -1.487 | -1.308 | -3.011 |
| 19 | 1.042 | -1.983 | -0.022 | -3.011 |
| 20 | -1.157 | -1.983 | 1.24 | -2.37 |
| 21 | -0.909 | -1.983 | 1.187 | -1.704 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.11211 |
| 0.0005 | 4.16771 |
| 0.0001 | 6.41271 |