| Motif | ZNF813.H13CORE.0.SG.A |
| Gene (human) | ZNF813 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 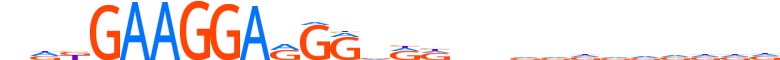 |
| LOGO (reverse complement) | 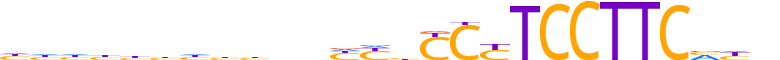 |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZNF813.H13CORE.0.SG.A |
| Gene (human) | ZNF813 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 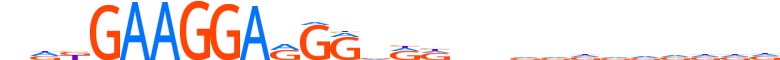 |
| LOGO (reverse complement) | 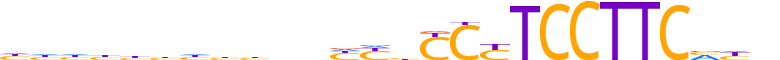 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 26 |
| Consensus | ndbGAAGGARGRdRdndddddddddd |
| GC content | 56.51% |
| Information content (bits; total / per base) | 17.835 / 0.686 |
| Data sources | HT-SELEX + Genomic HT-SELEX |
| Aligned words | 2264 |
| Previous names |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 0.982 | 0.97 | 0.958 | 0.941 | 0.865 | 0.855 |
| best | 0.982 | 0.97 | 0.96 | 0.943 | 0.889 | 0.873 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 338.559 | 0.964 | 0.979 | 0.934 |
| best | 444.387 | 0.984 | 0.985 | 0.941 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF813-like {2.3.3.72} (TFClass) |
| TFClass ID | TFClass: 2.3.3.72.1 |
| HGNC | HGNC:33257 |
| MGI | |
| EntrezGene (human) | GeneID:126017 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN813_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q6ZN06 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| Genomic HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZNF813.H13CORE.0.SG.A.pcm |
| PWM | ZNF813.H13CORE.0.SG.A.pwm |
| PFM | ZNF813.H13CORE.0.SG.A.pfm |
| Threshold to P-value map | ZNF813.H13CORE.0.SG.A.thr |
| Motif in other formats | |
| JASPAR format | ZNF813.H13CORE.0.SG.A_jaspar_format.txt |
| MEME format | ZNF813.H13CORE.0.SG.A_meme_format.meme |
| Transfac format | ZNF813.H13CORE.0.SG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 613.25 | 470.25 | 743.25 | 437.25 |
| 02 | 941.0 | 148.0 | 945.0 | 230.0 |
| 03 | 220.0 | 251.0 | 646.0 | 1147.0 |
| 04 | 9.0 | 10.0 | 2229.0 | 16.0 |
| 05 | 2248.0 | 1.0 | 10.0 | 5.0 |
| 06 | 2251.0 | 1.0 | 7.0 | 5.0 |
| 07 | 0.0 | 2.0 | 2258.0 | 4.0 |
| 08 | 30.0 | 1.0 | 2233.0 | 0.0 |
| 09 | 2221.0 | 8.0 | 12.0 | 23.0 |
| 10 | 722.0 | 108.0 | 1310.0 | 124.0 |
| 11 | 182.0 | 40.0 | 1990.0 | 52.0 |
| 12 | 290.0 | 91.0 | 1794.0 | 89.0 |
| 13 | 841.0 | 324.0 | 749.0 | 350.0 |
| 14 | 408.0 | 166.0 | 1425.0 | 265.0 |
| 15 | 406.0 | 214.0 | 1328.0 | 316.0 |
| 16 | 510.0 | 476.0 | 824.0 | 454.0 |
| 17 | 669.0 | 366.0 | 832.0 | 397.0 |
| 18 | 641.0 | 273.0 | 951.0 | 399.0 |
| 19 | 582.0 | 288.0 | 963.0 | 431.0 |
| 20 | 581.0 | 235.0 | 1046.0 | 402.0 |
| 21 | 634.0 | 256.0 | 957.0 | 417.0 |
| 22 | 691.0 | 258.0 | 942.0 | 373.0 |
| 23 | 615.0 | 239.0 | 1028.0 | 382.0 |
| 24 | 604.0 | 241.0 | 1050.0 | 369.0 |
| 25 | 685.0 | 212.0 | 1021.0 | 346.0 |
| 26 | 656.0 | 215.0 | 1077.0 | 316.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.271 | 0.208 | 0.328 | 0.193 |
| 02 | 0.416 | 0.065 | 0.417 | 0.102 |
| 03 | 0.097 | 0.111 | 0.285 | 0.507 |
| 04 | 0.004 | 0.004 | 0.985 | 0.007 |
| 05 | 0.993 | 0.0 | 0.004 | 0.002 |
| 06 | 0.994 | 0.0 | 0.003 | 0.002 |
| 07 | 0.0 | 0.001 | 0.997 | 0.002 |
| 08 | 0.013 | 0.0 | 0.986 | 0.0 |
| 09 | 0.981 | 0.004 | 0.005 | 0.01 |
| 10 | 0.319 | 0.048 | 0.579 | 0.055 |
| 11 | 0.08 | 0.018 | 0.879 | 0.023 |
| 12 | 0.128 | 0.04 | 0.792 | 0.039 |
| 13 | 0.371 | 0.143 | 0.331 | 0.155 |
| 14 | 0.18 | 0.073 | 0.629 | 0.117 |
| 15 | 0.179 | 0.095 | 0.587 | 0.14 |
| 16 | 0.225 | 0.21 | 0.364 | 0.201 |
| 17 | 0.295 | 0.162 | 0.367 | 0.175 |
| 18 | 0.283 | 0.121 | 0.42 | 0.176 |
| 19 | 0.257 | 0.127 | 0.425 | 0.19 |
| 20 | 0.257 | 0.104 | 0.462 | 0.178 |
| 21 | 0.28 | 0.113 | 0.423 | 0.184 |
| 22 | 0.305 | 0.114 | 0.416 | 0.165 |
| 23 | 0.272 | 0.106 | 0.454 | 0.169 |
| 24 | 0.267 | 0.106 | 0.464 | 0.163 |
| 25 | 0.303 | 0.094 | 0.451 | 0.153 |
| 26 | 0.29 | 0.095 | 0.476 | 0.14 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.08 | -0.185 | 0.272 | -0.257 |
| 02 | 0.507 | -1.332 | 0.511 | -0.896 |
| 03 | -0.94 | -0.809 | 0.132 | 0.705 |
| 04 | -3.95 | -3.863 | 1.368 | -3.455 |
| 05 | 1.377 | -5.267 | -3.863 | -4.406 |
| 06 | 1.378 | -5.267 | -4.152 | -4.406 |
| 07 | -5.684 | -4.973 | 1.381 | -4.562 |
| 08 | -2.878 | -5.267 | 1.37 | -5.684 |
| 09 | 1.365 | -4.046 | -3.708 | -3.126 |
| 10 | 0.243 | -1.642 | 0.837 | -1.506 |
| 11 | -1.127 | -2.606 | 1.255 | -2.354 |
| 12 | -0.665 | -1.81 | 1.151 | -1.832 |
| 13 | 0.395 | -0.555 | 0.279 | -0.479 |
| 14 | -0.326 | -1.218 | 0.921 | -0.755 |
| 15 | -0.331 | -0.967 | 0.851 | -0.58 |
| 16 | -0.104 | -0.173 | 0.375 | -0.22 |
| 17 | 0.167 | -0.434 | 0.384 | -0.353 |
| 18 | 0.124 | -0.725 | 0.518 | -0.348 |
| 19 | 0.028 | -0.672 | 0.53 | -0.271 |
| 20 | 0.026 | -0.874 | 0.613 | -0.341 |
| 21 | 0.113 | -0.789 | 0.524 | -0.304 |
| 22 | 0.199 | -0.782 | 0.508 | -0.415 |
| 23 | 0.083 | -0.857 | 0.595 | -0.392 |
| 24 | 0.065 | -0.849 | 0.616 | -0.426 |
| 25 | 0.19 | -0.976 | 0.588 | -0.49 |
| 26 | 0.147 | -0.962 | 0.642 | -0.58 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.82356 |
| 0.0005 | 2.30226 |
| 0.0001 | 5.38781 |