| Motif | ZNF814.H13INVITRO.0.PSG.A |
| Gene (human) | ZNF814 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 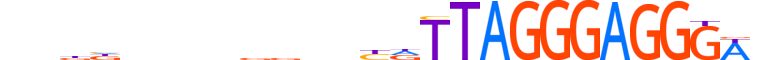 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZNF814.H13INVITRO.0.PSG.A |
| Gene (human) | ZNF814 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 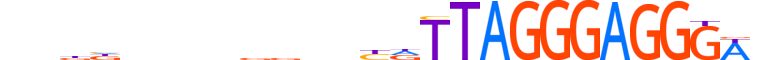 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 26 |
| Consensus | ndddnnnnddnnbdTTAGGGAGGGWn |
| GC content | 54.03% |
| Information content (bits; total / per base) | 21.19 / 0.815 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX |
| Aligned words | 478 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 1 (1) | 0.591 | 0.591 | 0.524 | 0.524 | 0.441 | 0.441 | 11.06 | 11.06 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 1 experiments | median | 0.999 | 0.999 | 0.992 | 0.99 | 0.882 | 0.892 |
| best | 0.999 | 0.999 | 0.992 | 0.99 | 0.882 | 0.892 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 62.52 | 0.386 | 0.532 | 0.471 |
| best | 232.06 | 0.575 | 0.638 | 0.634 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Multiple dispersed zinc fingers {2.3.4} (TFClass) |
| TF subfamily | ZNF417-like {2.3.4.1} (TFClass) |
| TFClass ID | TFClass: 2.3.4.1.4 |
| HGNC | HGNC:33258 |
| MGI | |
| EntrezGene (human) | GeneID:730051 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN814_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | B7Z6K7 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 1 overall: 0 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZNF814.H13INVITRO.0.PSG.A.pcm |
| PWM | ZNF814.H13INVITRO.0.PSG.A.pwm |
| PFM | ZNF814.H13INVITRO.0.PSG.A.pfm |
| Threshold to P-value map | ZNF814.H13INVITRO.0.PSG.A.thr |
| Motif in other formats | |
| JASPAR format | ZNF814.H13INVITRO.0.PSG.A_jaspar_format.txt |
| MEME format | ZNF814.H13INVITRO.0.PSG.A_meme_format.meme |
| Transfac format | ZNF814.H13INVITRO.0.PSG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 151.0 | 57.0 | 141.0 | 129.0 |
| 02 | 129.75 | 61.75 | 110.75 | 175.75 |
| 03 | 84.0 | 51.0 | 185.0 | 158.0 |
| 04 | 72.0 | 59.0 | 255.0 | 92.0 |
| 05 | 107.0 | 97.0 | 190.0 | 84.0 |
| 06 | 123.0 | 94.0 | 161.0 | 100.0 |
| 07 | 135.0 | 71.0 | 137.0 | 135.0 |
| 08 | 87.0 | 100.0 | 167.0 | 124.0 |
| 09 | 82.0 | 77.0 | 223.0 | 96.0 |
| 10 | 88.0 | 67.0 | 199.0 | 124.0 |
| 11 | 109.0 | 155.0 | 107.0 | 107.0 |
| 12 | 72.0 | 151.0 | 154.0 | 101.0 |
| 13 | 36.0 | 237.0 | 38.0 | 167.0 |
| 14 | 185.0 | 12.0 | 198.0 | 83.0 |
| 15 | 1.0 | 41.0 | 6.0 | 430.0 |
| 16 | 0.0 | 0.0 | 4.0 | 474.0 |
| 17 | 477.0 | 0.0 | 0.0 | 1.0 |
| 18 | 0.0 | 1.0 | 476.0 | 1.0 |
| 19 | 0.0 | 0.0 | 478.0 | 0.0 |
| 20 | 0.0 | 0.0 | 478.0 | 0.0 |
| 21 | 475.0 | 1.0 | 1.0 | 1.0 |
| 22 | 2.0 | 1.0 | 472.0 | 3.0 |
| 23 | 0.0 | 2.0 | 476.0 | 0.0 |
| 24 | 4.0 | 1.0 | 408.0 | 65.0 |
| 25 | 343.75 | 26.75 | 25.75 | 81.75 |
| 26 | 129.5 | 95.5 | 165.5 | 87.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.316 | 0.119 | 0.295 | 0.27 |
| 02 | 0.271 | 0.129 | 0.232 | 0.368 |
| 03 | 0.176 | 0.107 | 0.387 | 0.331 |
| 04 | 0.151 | 0.123 | 0.533 | 0.192 |
| 05 | 0.224 | 0.203 | 0.397 | 0.176 |
| 06 | 0.257 | 0.197 | 0.337 | 0.209 |
| 07 | 0.282 | 0.149 | 0.287 | 0.282 |
| 08 | 0.182 | 0.209 | 0.349 | 0.259 |
| 09 | 0.172 | 0.161 | 0.467 | 0.201 |
| 10 | 0.184 | 0.14 | 0.416 | 0.259 |
| 11 | 0.228 | 0.324 | 0.224 | 0.224 |
| 12 | 0.151 | 0.316 | 0.322 | 0.211 |
| 13 | 0.075 | 0.496 | 0.079 | 0.349 |
| 14 | 0.387 | 0.025 | 0.414 | 0.174 |
| 15 | 0.002 | 0.086 | 0.013 | 0.9 |
| 16 | 0.0 | 0.0 | 0.008 | 0.992 |
| 17 | 0.998 | 0.0 | 0.0 | 0.002 |
| 18 | 0.0 | 0.002 | 0.996 | 0.002 |
| 19 | 0.0 | 0.0 | 1.0 | 0.0 |
| 20 | 0.0 | 0.0 | 1.0 | 0.0 |
| 21 | 0.994 | 0.002 | 0.002 | 0.002 |
| 22 | 0.004 | 0.002 | 0.987 | 0.006 |
| 23 | 0.0 | 0.004 | 0.996 | 0.0 |
| 24 | 0.008 | 0.002 | 0.854 | 0.136 |
| 25 | 0.719 | 0.056 | 0.054 | 0.171 |
| 26 | 0.271 | 0.2 | 0.346 | 0.183 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.231 | -0.726 | 0.163 | 0.076 |
| 02 | 0.081 | -0.648 | -0.075 | 0.382 |
| 03 | -0.347 | -0.835 | 0.433 | 0.276 |
| 04 | -0.498 | -0.693 | 0.751 | -0.258 |
| 05 | -0.109 | -0.206 | 0.459 | -0.347 |
| 06 | 0.029 | -0.237 | 0.295 | -0.176 |
| 07 | 0.12 | -0.512 | 0.135 | 0.12 |
| 08 | -0.313 | -0.176 | 0.331 | 0.037 |
| 09 | -0.371 | -0.433 | 0.618 | -0.216 |
| 10 | -0.301 | -0.569 | 0.505 | 0.037 |
| 11 | -0.091 | 0.257 | -0.109 | -0.109 |
| 12 | -0.498 | 0.231 | 0.251 | -0.166 |
| 13 | -1.171 | 0.678 | -1.119 | 0.331 |
| 14 | 0.433 | -2.19 | 0.5 | -0.359 |
| 15 | -3.863 | -1.046 | -2.776 | 1.271 |
| 16 | -4.363 | -4.363 | -3.084 | 1.368 |
| 17 | 1.375 | -4.363 | -4.363 | -3.863 |
| 18 | -4.363 | -3.863 | 1.373 | -3.863 |
| 19 | -4.363 | -4.363 | 1.377 | -4.363 |
| 20 | -4.363 | -4.363 | 1.377 | -4.363 |
| 21 | 1.37 | -3.863 | -3.863 | -3.863 |
| 22 | -3.531 | -3.863 | 1.364 | -3.283 |
| 23 | -4.363 | -3.531 | 1.373 | -4.363 |
| 24 | -3.084 | -3.863 | 1.219 | -0.598 |
| 25 | 1.048 | -1.454 | -1.49 | -0.374 |
| 26 | 0.079 | -0.221 | 0.322 | -0.307 |
| P-value | Threshold |
|---|---|
| 0.001 | -1.53674 |
| 0.0005 | 0.12866 |
| 0.0001 | 3.60561 |